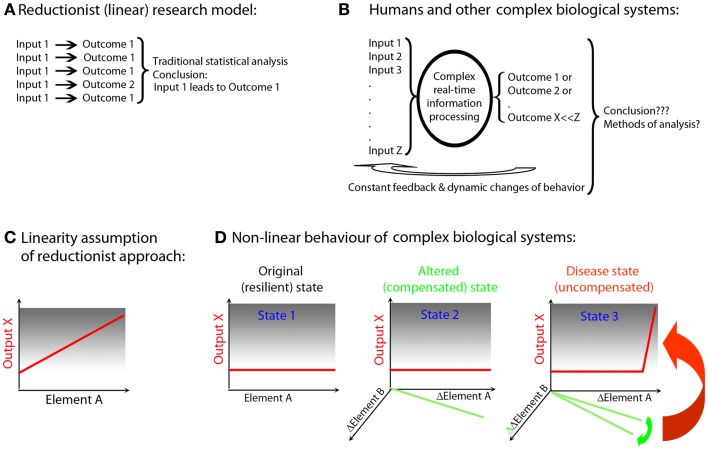
Figure 1.
Simplified animal models versus complex biological systems exemplified by human polygenic diseases. (A) Reductionist (linear) research model (e.g., experimental autoimmune encephalomyelitis): current animal studies are almost exclusively performed in a single animal species of a single genetic strain (usually the one that is susceptible to induction of the disease). Furthermore, the animals are housed in the same (often pathogen-free) environment; they are exposed to identical food and identical environmental stimuli, which leads to synchronization of circadian rhythms, similar levels of activity, etc. Disease is induced by identical regimens applied in a highly synchronized manner to animals of the same age and often only of single sex. Therefore, animal experiments utilize highly simplified input (input 1). Despite standardized input, the outcome is usually somewhat heterogeneous (outcome 1 and 2), but an application of traditional statistical methods leads to clear conclusions. These conclusions are often readily generalized across species and across diverse environmental inputs and disease triggers. (B) Humans and other non-artificial complex biological systems: measurements in the complex biological system exemplified by a human being affected by a disease are the results of multiple different inputs (i.e., an outbred genetic background, many environmental influences such as type, dose, and virulence of an infectious agent, diverse food, premorbidities, and drug regimens influencing both the metabolome and the microbiome, varied endocrine regulations resulting from circadian and reproductive rhythms and aging). The organism processes these varied inputs, utilizing complex decision-making mechanisms and the outcomes are also diverse (e.g., maintenance of heath or development of the disease of varied severity). Furthermore, the outcomes are processed by the organism as additional inputs through constant ubiquitous feedback loops, leading to dynamic changes of behavior. Current statistical methods are largely inadequate for analysis of such complex datasets. As a consequence, frequently no reproducible conclusions are reached. (C) Linearity assumption of reductionist approach: reductionist research methods are based on assumption that if an element (e.g., gene, its transcript or protein; Element A) is linked to a disease process (output X), then an observed or induced change in the element has to be reflected in the change in the output. (D) Non-linear behavior of biological systems: the existence of functional networks consisting of interacting elements with partially overlapping functions allows biological systems to retain a given output in a normal (non-disease) state if only one or few elements have been altered (this is called robustness of the system). For example, an alteration in Element A was compensated by a reciprocal change in Element B (State 2). However, despite maintenance of the same output, this new state is different from the original state (healthy; State 1), even though phenotypically there is no evidence of a disease. Such an altered state of the system is more fragile, insofar as any subsequent change in Element B may now cause a dramatic change in the output X (i.e., disease state; State 3).