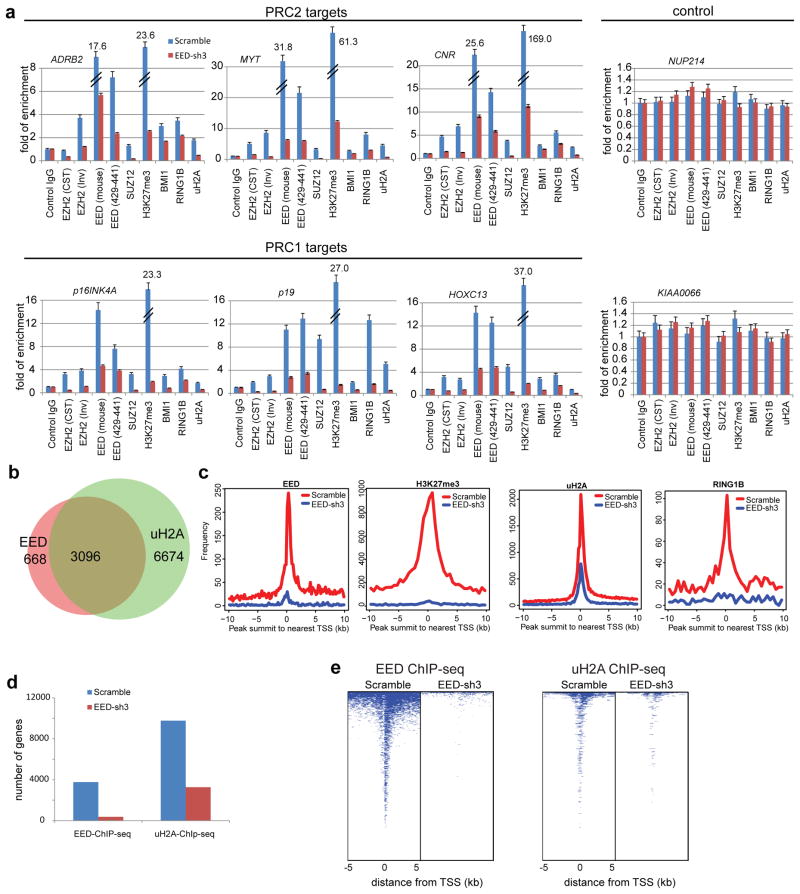
Figure 5. EED is critical for PRC1 recruitment to target genes.
a, ChIP-qPCR analysis of indicated proteins at PRC2 target genes ADRB2, MYT1 and CNR and PRC1 target genes p16INK4A/CDKN2A, p19/CDKN2D and HOXC13. NUP214 and KIAA0066 were used as negative controls. Antibodies used for ChIP: mouse control IgG, rabbit control IgG, anti-EZH2 (CST) (Cell Signaling, cat# 5246), anti-EZH2 (Inv) (Invitrogen, cat# 49-1043), anti-EED (Millipore, cat# 05-1320), anti-EED (aa429-441) (Millipore, cat# 09-774), anti-SUZ12, anti-BMI1, anti-RING1B, anti-H3K27me3 and anti-uH2A. ChIP assays were biologically repeated twice and qPCR analyses were done in triplicate. All error bars represent ±s.e.m. b, Venn diagrams show that 82.3% of EED bound genes in DU145 cells are also enriched with uH2A. c, Knock-down of EED attenuates the occupancy of EED, uH2A, H3K27me3 and RING1B at PcG target promoters. Genome-wide pattern of uH2A modification, H3K27me3, EED and RING1B in DU145 scramble control cells (red) and DU145 EED-sh3 cells (blue) are noted at all annotated gene promoters. d, Bar graph of genes with EED occupancy (left panel) and uH2A (right panel) modification in scramble or EED-sh3 cells. e, Heatmap representation of genomic regions with binding profiles of EED (left panel) and uH2A (right panel) in DU145 scramble or EED-sh3 cells 5kb flanking the transcriptional start site of genes.