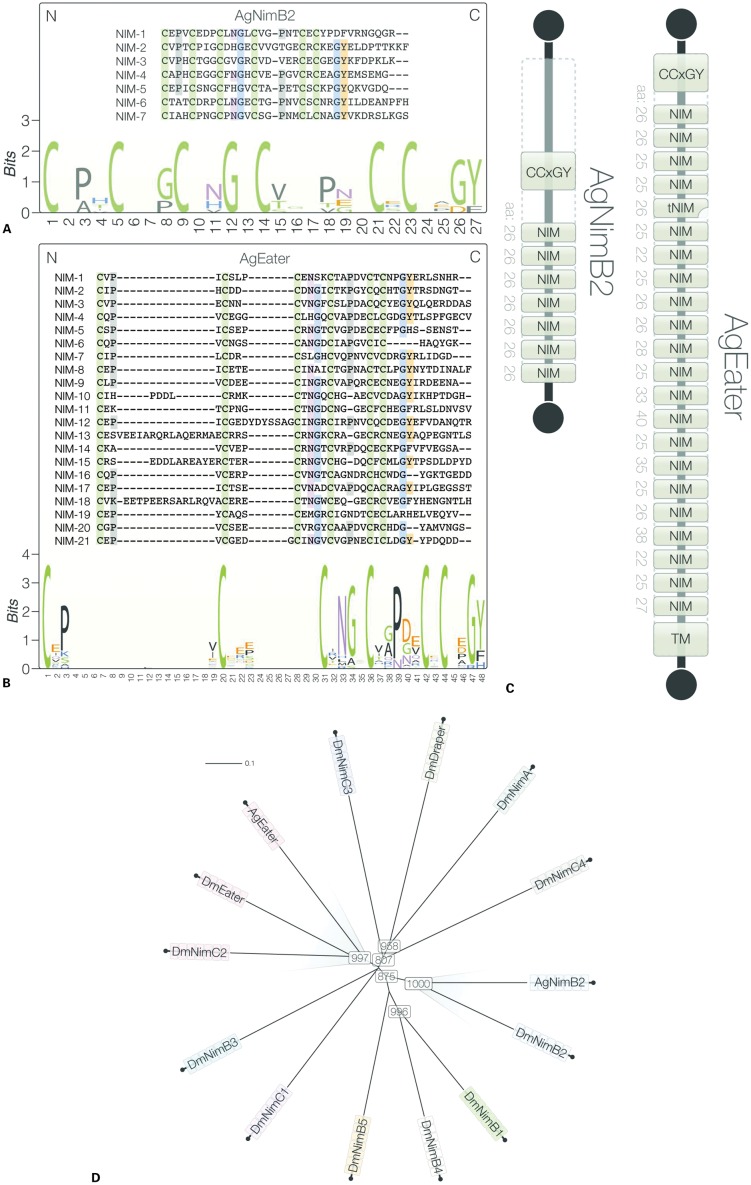
Figure 1.
Bioinformatic analysis of AgNimB2 and AgEater (A and B) ClustalW sequence alignment of predicted NIM repeats in AgNimB2 and AgEater with the consensus amino acids highlighted. Both alignments were used to produce a HMM logo representing the conversed amino acids. (C) Diagrammatic representation of the AgNimB2 and AgEater proteins, showing the N- (top) and C-termini (bottom), the transmembrane domain (TM), and NIM repeats. The amino acid length is indicated to the left of each repeat. A truncated NIM repeat in AgEater (tNIM) is represented by a notched rectangle. (D) Unrooted tree generated from the Nimrod superfamily members of Drosophila melanogaster, AgNimB2, and AgEater. Orthologs are highlighted in blue (NimB2) and red (Eater). Numbers represent bootstrap values.