Figure 5. Methylation of RUNX1 at R223 regulates its interaction with DPF2.
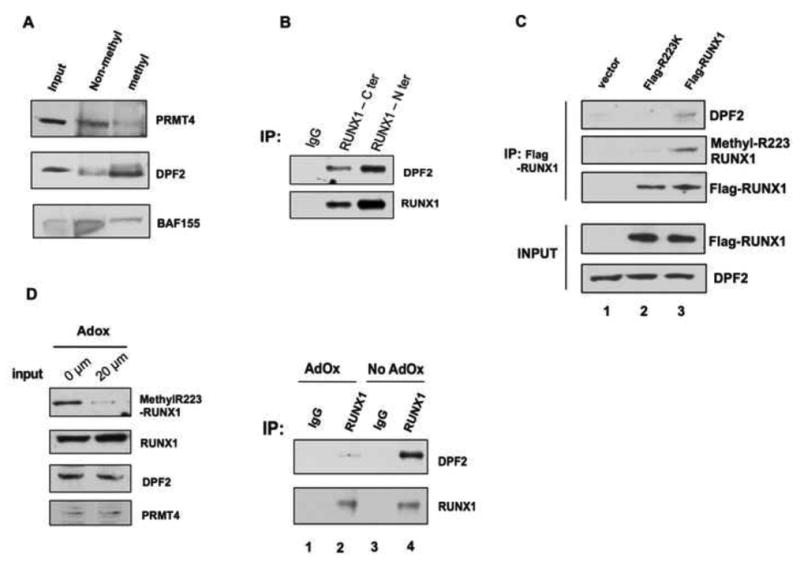
(A). DPF2 is preferably pulled down by a methylR223- RUNX1 peptide, while the unmodified R223-RUNX1 peptide interacts more strongly with PRMT4. BAF155 is used as a control, which shows no preference for binding to either the modified or the unmodified peptide. Input: ten percent of the used nuclear extract.
(B). The endogenous RUNX1 and DPF2 proteins physically interact in vivo. Two anti-RUNX1 antibodies were used to immunoprecipitate DPF2 from HEL cell nuclear extract. DPF2 was detected using an anti-DPF2 antibody. Pre-immune rabbit serum was used as control.
(C). The interaction between RUNX1 and DPF2 is dependent on the RUNX1 methylation status. Overexpressed Flag-RUNX1 is immunoprecipitated using an anti-Flag antibody. DPF2 is co-precipitated with Flag-RUNX1, but not the Flag-R223K mutant (compare lane 1 vs. lane 3).
(D). Treatment of cells with AdOx reduces the level of RUNX1 methylation (input), abrogating its interaction with DPF2 (compare lane 2 vs. lane 4).
See also Figure S5.
