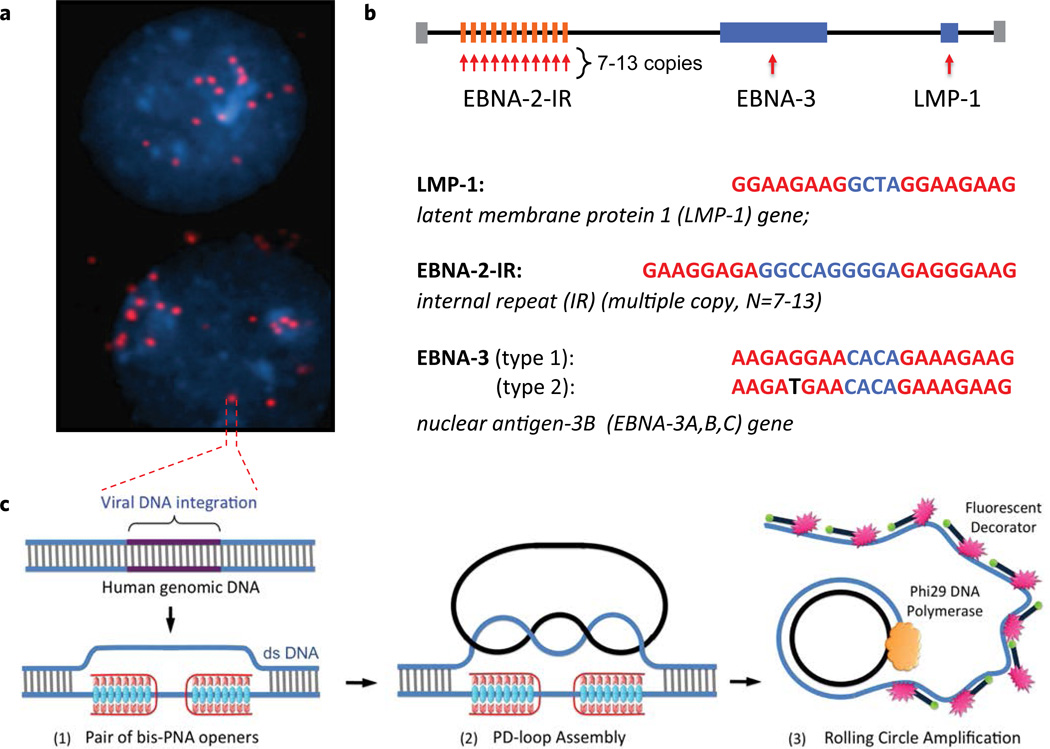
Figure 1.
The PNA-RCA method for detection of short specific DNA target site at the single cell. (a) Multiple fluorescent spots were observed in the BC-1 cells (EBV-positive) when probe corresponding to the LMP-1 gene encoding major transforming protein of Epstein-Barr Virus was applied. The fluorescent signals were acquired separately using two filter sets: DAPI for DNA and Cy3 for labeled RCA product. (b) Target sites in the EBV genomic DNA used in this study. Sequences targeted by PNAs are shown in red. EBNA-3(G)-EBV type 1 signature sites within the EBNA-3 gene differ by a single nucleotide (SNP) from EBNA-3(T)-EBV type 2 in the PNA binding sequence. Mismatch is shown in black. (c) Schematic depiction of the PNA-RCA method for sensitive and specific targeting of unique sequences on non-denatured human genomic DNA. (I) The PNA openers specifically bind to two closely located homopurine DNA sites that are separated by several mixed purine-pyrimidine bases and locally open the double-stranded DNA. (II) The opened region serves as a target for hybridization and ligation of an oligonucleotide probe to form a PD-loop. (III) The small circle on duplex DNA serves as a template for isothermal RCA reaction, which yields a long, single-stranded amplicon that contains thousands of copies of the target sequence. For the fluorescence-based detection, a fluorophore-tagged decorator probes are hybridized to the RCA product.