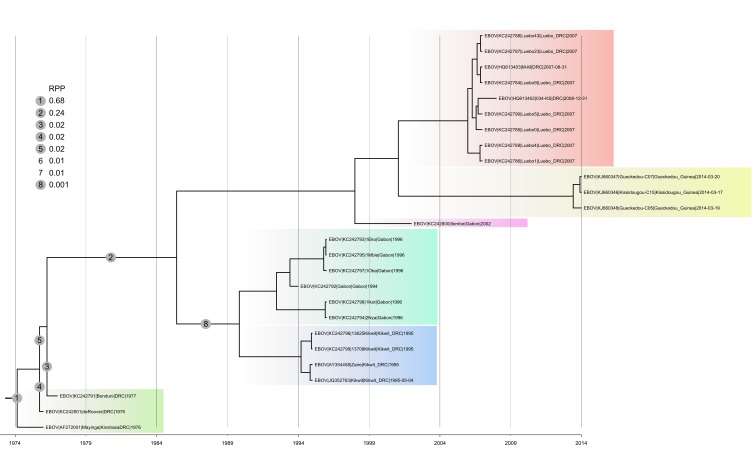
Maximum clade credibility tree of concatenated coding sequences belonging to the Zaïre clade.
MCC tree and branch root posterior probabilities (RPP) derived from the analysis run under a constant population size model (the two other models ended up with generating very similar results) and an uncorrelated relaxed clock (lognormal). In the top left corner the complete list of branches that appeared at least once in the posterior tree sample and the according RPP. Note that two possible root locations (6 and 7) do not appear in the tree as the MCC tree did not comprise the corresponding branches. All internal branches linking coloured clades/groups received very good support (posterior probability: 1.00). The only exception was the branch defining the clade comprising Guinea 2014 EBOV and DRC 2007/2008 EBOV, which was only moderately supported (posterior probability comprised between 0.56 and 0.68).