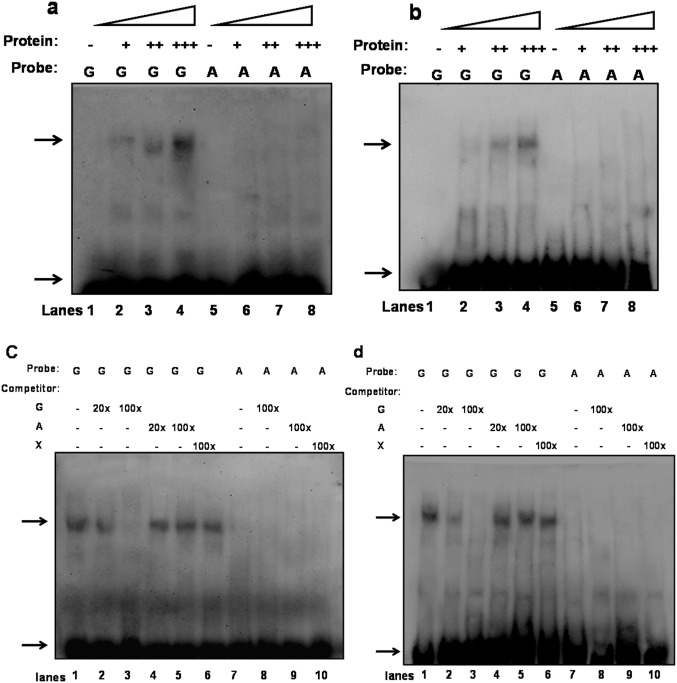
Figure 3. EMSA was performed to quantitatively analyse DNA-protein interaction.
All samples contained binding buffer (1.5 µL), Poly dI:dC (1.5 µL), G allele or A allele probe (0.3 pmol), and nuclear protein extracts (5–15 µg). Band designations: upper arrow, protein-DNA probe complex; lower arrow, free DNA monocyte-derived macrophages (a) or THP-1 cell (b) nuclear protein bound to labelled DNA probe, and lanes 1–4 and lanes 5–8 contained 0, 5, 9, and 15 µg protein, respectively. Unlabelled cold probe competitively bound monocyte-derived macrophages (c) and THP-1 cell (d) nuclear protein. Specificity of binding was tested by competition with a 20- or 100-fold molar excess of unlabelled DNA probe added to the reaction mixture. Lanes 1 and 7: No competitor; lanes 2, 3, 9: self-competitor; lanes 4–6, 8, 10: cross-competitor. The results are representative of several independent experiments. Probe “G” for rs57137919G and “A” for rs57137919A biotin-labelled probes. Competitor “G” for rs57137919G, “A” for rs57137919A, and “X” for non-specific unlabelled probes.