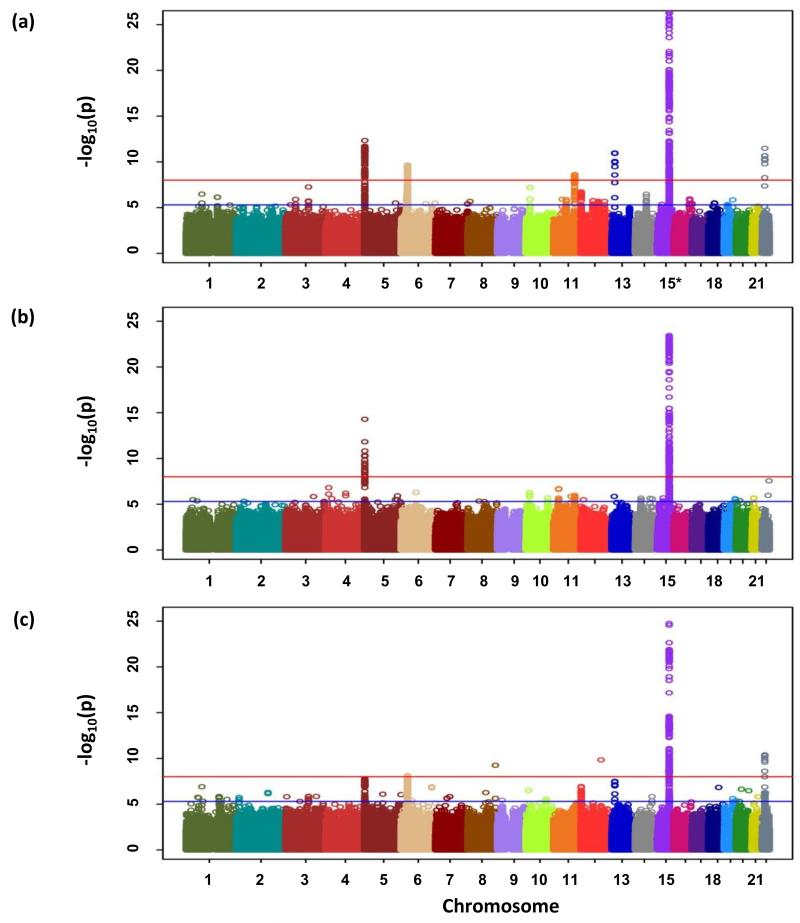
Figure 1. Genome-wide P-values (−log10P, y axis) plotted against their respective chromosomal positions (x axis).
(a) All lung cancer, (b) AD and (c) SQ. Shown are the genomewide P-values (two-sided) obtained using the Cochran-Armitage trend test from analysis of 8.9 million successfully imputed autosomal SNPs in 11,348 cases and 15,861 controls from discovery phase. The red and blue horizontal lines represent the significance threshold of P=5.0×10−8 and P=5.0×10−6 respectively. Any region contains at least one association signal better than P=5.0×10−6 were selected for the in silico replication.