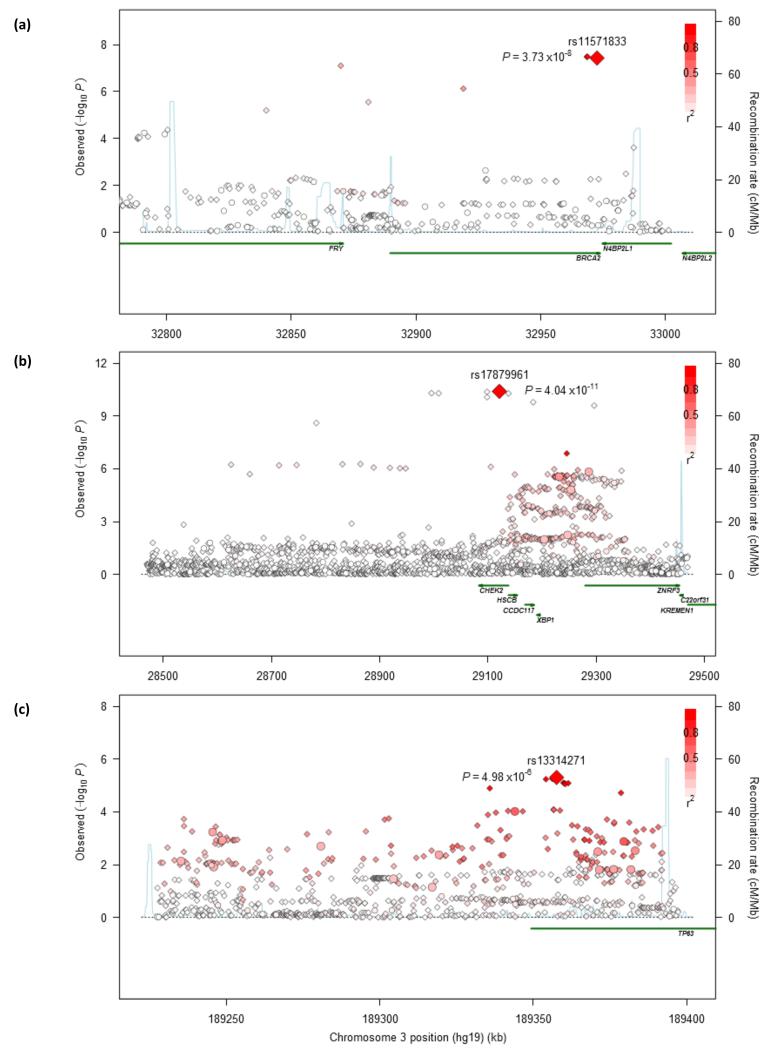
Figure 3. Regional plots of association results and recombination rates for the 13q13.1 in SQ (a), 22q12.1 in SQ (b) and 3q28 susceptibility loci in AD (c).
SQ related panels (a, b) were based on 3,275 SQ and 15,038 controls from discovery phase; and AD related panel (c) was based on 3,442 AD and 14,894 controls from discovery phase. Association results of both genotyped (circles) and imputed (diamonds) SNPs in the GWAS samples and recombination rates for each locus: For each plot, −log10P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top genotyped SNP in each combined analysis is a large diamond and is labeled by its rsID. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2=0) through to dark red (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale.