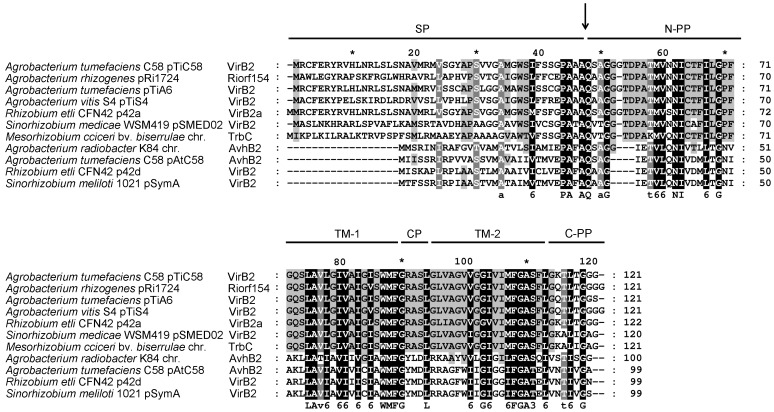
Figure 1. Amino acid sequence alignment of VirB2 family proteins.
Multiple amino acid sequence alignment of VirB2 homologues with ClustalW2 [50]. The organism/plasmid name for each homolog is indicated on the left of the aligned sequence. UniProt accession numbers: Agrobacterium tumefaciens pTiC58, P17792; Agrobacterium rhizogenes pRi1724, Q9F5A1; Agrobacterium tumefaciens pTiA6, P05351; Agrobacterium vitis pTiS4, B9K417; Rhizobium etli p42a, Q2K2L1; Sinorhizobium medicae pSMED02; A6UMA7, Mesorhizobium ciceri chromosome (chr.), E8TGI0; Agrobacterium radiobacter K84 chromosome (chr.), B9JE70; Agrobacterium tumefaciens pAtC58, Q7D3S1; Rhizobium etli p42d, Q8KIM6; Sinorhizobium meliloti pSymA, Q92YZ4. The amino acid residues identical in all proteins are in black and those conserved in most but not all of the proteins are in gray. The arrow indicates the processing site of VirB2 encoded by pTiC58. Each region/domain of VirB2 is indicated: SP, signal peptide; TM-1, trans-membrane domain 1; CP, cytoplasmic domain; TM-2, trans-membrane domain 2; N-PP, N-terminal periplasmic domain; C-PP, C-terminal periplasmic domain.