Fig. 5.
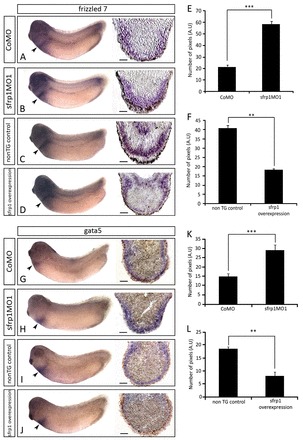
Experimental sfrp1 manipulation causes complementary effects on non-myocardial marker gene expression. (A-D) WISH analysis showing frizzled 7 (fzd7) expression (arrowheads) in CoMO-injected (A), sfrp1MO1-injected (B), non-transgenic control (C) and sfrp1-overexpressing transgenic (D) embryos, together with corresponding sections through the posterior pericardium. (G-J) WISH analysis showing gata5 expression (arrowheads) in CoMO (G), sfrp1MO1 (H), non-transgenic control (I) and sfrp1-overexpressing (J) embryos, with corresponding posterior pericardial sections. (E,F,K,L) Analysis of fzd7 (E,F) and gata5 (K,L) sections representing the number of pixels measured from the apex of the section to the edge of gene expression staining. CoMO compared with sfrp1MO1: n=4, ***P<0.001 (E,K). Non-transgenic control compared with sfrp1 overexpression: n=3, **P<0.01 (F,L). Data are mean ± s.e.m.
