Fig. 8.
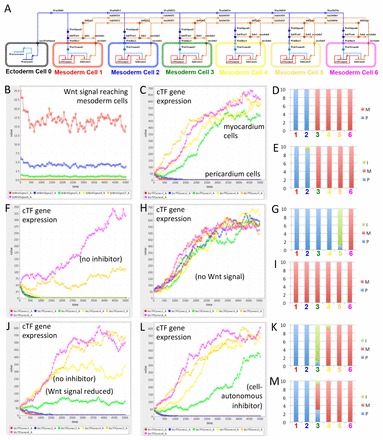
One-dimensional multicellular model of a Wnt negative-feedback gene regulatory network controlling myocardium differentiation. (A) One-dimensional model of a gene regulatory network (GRN) designed in BioTapestry Editor. Note the ectodermal cell (source of Wnt signal) at the far left, with distal to medial cardiac mesoderm cells from left to right, in which myocardium differentiation potential is controlled by cardiogenic transcription factor (cTF) gene expression. Colours of cells correspond to simulation output from the Dizzy simulator (see below). (B) Simulated value of Wnt signal (in arbitrary units, but related to amount) reaching cardiac mesoderm cells. (C) Simulated expression of cTF gene expression (in arbitrary units related to amount) in different cardiac mesoderm cells. Note that simulation of the full model (as illustrated in A) predicts clear patterning into two populations of cells according to cTF gene expression: into prospective non-muscular cells that have lost cTF expression (here simply called pericardium) and prospective muscular cells gaining strong cTF expression (myocardium). (D) Repeated simulation shows reproducibility of output suggesting reliability of patterning. Ten random simulations are illustrated in a stacked column chart. Cells were attributed to the pericardium (P) if the cTF expression was lower than 30 and to the myocardium (M) if higher than 300. Coloured numbers correspond to cells illustrated in A and simulation output in B and C. (E) Simulation of a GRN lacking negative regulation of inhibitor gene expression (i.e. inhibitor gene expression only regulated by cTF genes) suggests still relatively reliable patterning with only occasional cells with predicted intermediate (I) (i.e. between 30 and 300) values of cTF expression. (F) Simulation of an inhibitor loss-of-function experiment predicting many fewer cells with sufficient cTF gene expression for myocardium differentiation. (G) Repeated simulation of experiment as in F. (H) Simulation of Wnt signal loss-of-function experiment (or Wnt and inhibitor double loss-of-function experiment) confirming that the GRN is designed with the default for cardiac mesoderm cells being myocardium differentiation. (I) Repeated simulation of experiment as in H. (J) Simulation of an alternative GRN with only Wnt regulation (i.e. no inhibitor) but with values for Wnt signal adjusted so that the simulation usually predicts at least two pericardial and two myocardial cells suggests less clear patterning giving rise to cells with intermediate identity. (K) Repeated simulation of the alternative GRN as in J. (L) Simulation of an alternative GRN with a cell-autonomous inhibitor predicts relatively clear patterning into pericardial and myocardial cells. (M) Repeated simulation of the alternative GRN as in L suggests less reliable patterning into two pericardium and four myocardium cells than the GRN illustrated in A (compare with D). See supplementary material Models 1-9.
