Fig. 1.
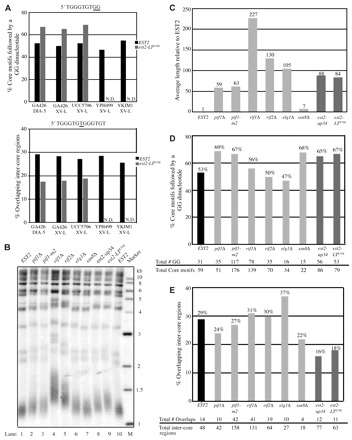
Disruption of negative telomere length regulation does not result in a specific telomere sequence phenotype. The indicated strains were grown for a minimum of 75 generations before cloning of telomeres by ligation-mediated PCR from the ADE2 marked chromosome V-R (DIA-5) or from chromosome XV-L. Chromosome V-R data for the EST2, est2-up34 and est2-LTE76K GA426 strains were previously published (Ji et al., 2008) and are included for comparison. (A) Telomere sequence characteristics of the indicated chromosome ends expressing either EST2 (black bar) or est2-LTE76K (gray bar). N.D., not determined. (B) Telomere length of GA426 strains determined by Southern blotting. Genomic DNA from the indicated strains was cleaved with XhoI, Southern blotted, and probed with a radiolabeled telomeric fragment. Marker sizes are in kilobases (kb). (C) Average length of 9–21 cloned telomeres used to generate data in D and E. Telomere length is normalized to the GA426 EST2 strain. Black bar, EST2; dark gray bars, est2-up34 and est2-LTE76K; light gray bars, six additional mutants. (D) Percentage of core motifs followed by a GG dinucleotide in telomeres from the indicated GA426 strains. Primary data for each strain are indicated (bottom). Gray-scale categories are the same as in C. (E) Percentage of inter-core region overlaps in the same telomeres analyzed in D. Primary data for each strain are indicated (bottom). Gray-scale categories are the same as in C and D.
