Figure 3.
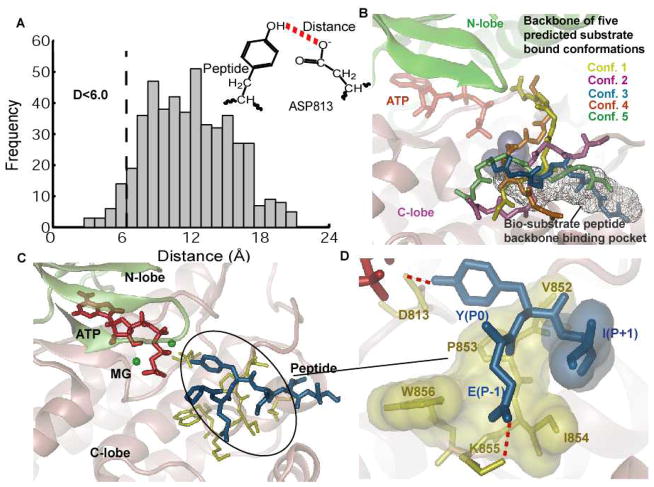
Results for docking the peptide Y1068 to the EGFR TKD. A) Histogram of the distance between the oxygen in hydroxyl group of the tyrosine and the oxygen atom in the carboxylate group of the D813 for all the 500 predicted conformations by ensemble docking. Conformations with the above distance less than 6 Å were clustered into 5 clusters based on their backbone RMSD are shown in panel B. B) Five predicted peptide clusters of Y1068 bound conformations generated by ensemble docking that satisfied the distance criteria in panel A. The bi-substrate peptide binding pocket is also shown in the background, for reference. All 5 conformations were re-ranked using the MMPBSA method (see Table 1). C) Bound conformation of the peptide Y1068 to EGFR TKD in the presence of ATP and two MG2+ ions after 10 ns of MD simulation. The ATP atoms are depicted in red, MG2+ in green, peptide in blue, and residues in EGFR TKD that interact with the peptide in yellow. D) Depiction of the main interactions between the peptide and the kinase: Y(P0) hydrogen-bonds with D813; E(P−1) forms a salt bridge interaction with residue K855; the hydrophobic residue I(P+1) interacts with a hydrophobic surface in EGFR TKD consisting of the residues V852-P853-I854-W856. All hydrophobic residues are depicted as transparent surfaces.