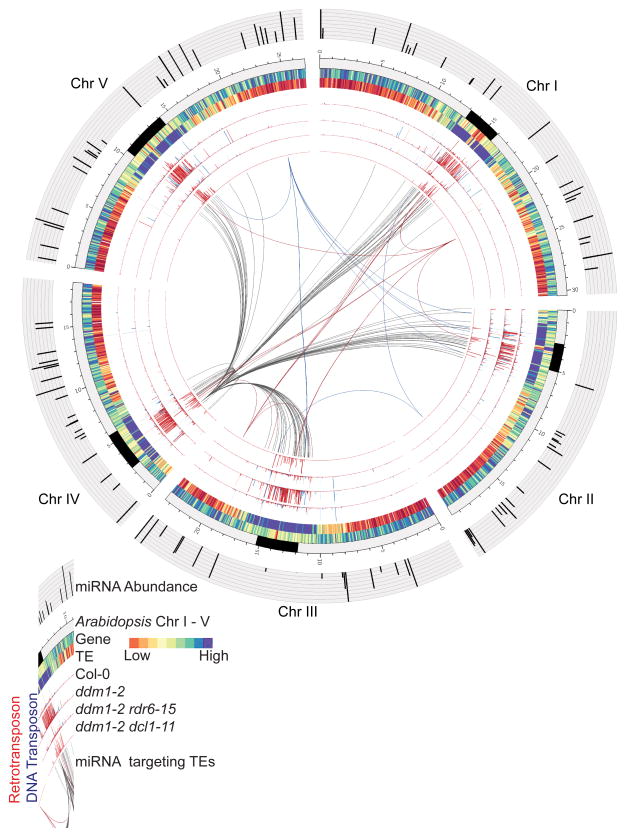
Figure 1. miRNAs trigger RDR6-dependent 21-nt epigenetically activated (ea)siRNA biogenesis from reactivated transposons.
Whole-genome representation illustrating miRNAs triggering widespread easiRNA biogenesis at transposons in Arabidopsis. Outermost to innermost tracks depicting: miRNA abundance in ddm1-2 (Histogram); Arabidopsis Chromosome I – V, pericentromeric region (black) (Ideogram); Gene and transposon frequency, low density (red), high density (blue) (Heat-map); Retrotransposon derived 21-nt sRNAs (dark red = unique, light red = multiple mapping) and DNA transposon-derived 21-nt sRNAs (dark blue = unique, light blue = multiple mapping) 21-nt siRNAs in order of Col-0, ddm1-2, ddm1-2 rdr6-15, and ddm1-2 dcl1-11 (Histogram); miRNAs targeting transposons (Connectors); miR859a (Chr I, red), miR390a (Chr II, blue), miR172d (Chr III, red), eamiR2 ATHILAIV (Chr IV, grey) and miR172e (Chr V, blue). Transposons are post-transcriptionally targeted by 50 known miRNAs and newly discovered eamiRNAs (Table 1) giving rise to abundant RDR6-dependent 21-nt easiRNAs at transposons in Arabidopsis. (Refer to Extended Data Fig. 1; Supplementary Table 1; 3).