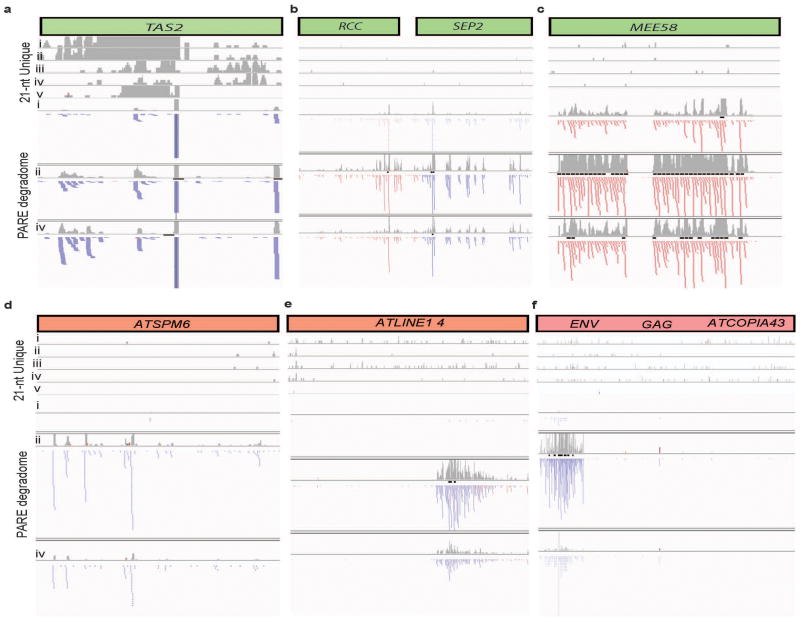
Extended Data Figure 2. miRNA target genes and transposons that do not promote tasiRNA nor easiRNA, respectively, have degradation covering the entire region.
Read pattern distribution of 21-nt unique reads (represented as a histogram of read density (grey bars)) and PARE signatures at (a) TAS2 (AT2G39681), (b) RCC (AT3G02300), SEP2 (AT3G02310), (c) MEE58 (AT4G13940), and transposons (d) ATENSPM6 (AT2G06720), (e) ATLINE1_4 (AT2G15540), (f) ATCOPIA43 (AT3G0410), in track order Col-0, ddm1-2, rdr6-15, ddm1-2 rdr6-15 and ddm1-2 dcl1-11.