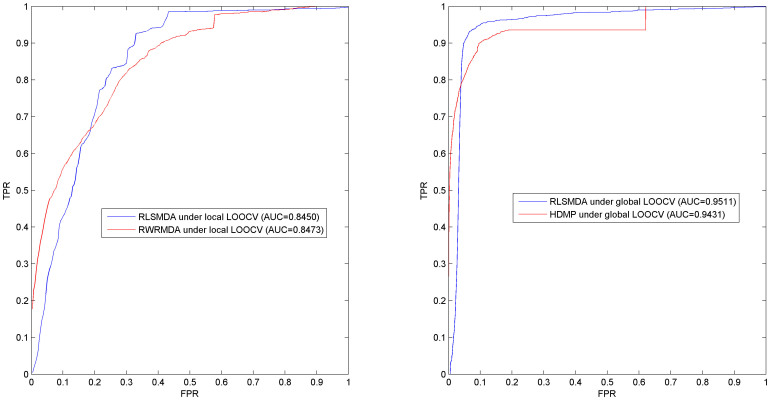
Figure 1. Method comparison: (left) Comparison between RLSMDA and RWRMDA proposed by Chen, et al.47 in terms of ROC curve and AUC based on local leave-one-out cross validation on 1394 known experimentally verified miRNA–disease associations.
RLSMDA obtained comparable performance in the local LOOCV as RWRMDA, while RWRMDA cannot predict disease-related miRNAs for diseases without known related miRNAs and all the diseases simultaneously. RLSMDA can successfully solve these two critical shortcomings of RWRMDA. (right) Comparison between RLSMDA and HDMP in the term of global LOOCV. RLSMDA and HDMP obtained the AUC of 0.9511 and 0.9431, respectively. Although only slight improvement has been obtained here, RLSMDA can predict the potential miRNAs for diseases which do not have known related miRNAs, which has solved the most critical limitation of HDMP. The performance of RLSMDA could be further improved by introducing the information of miRNA family and cluster as what has been done in the method of HDMP.