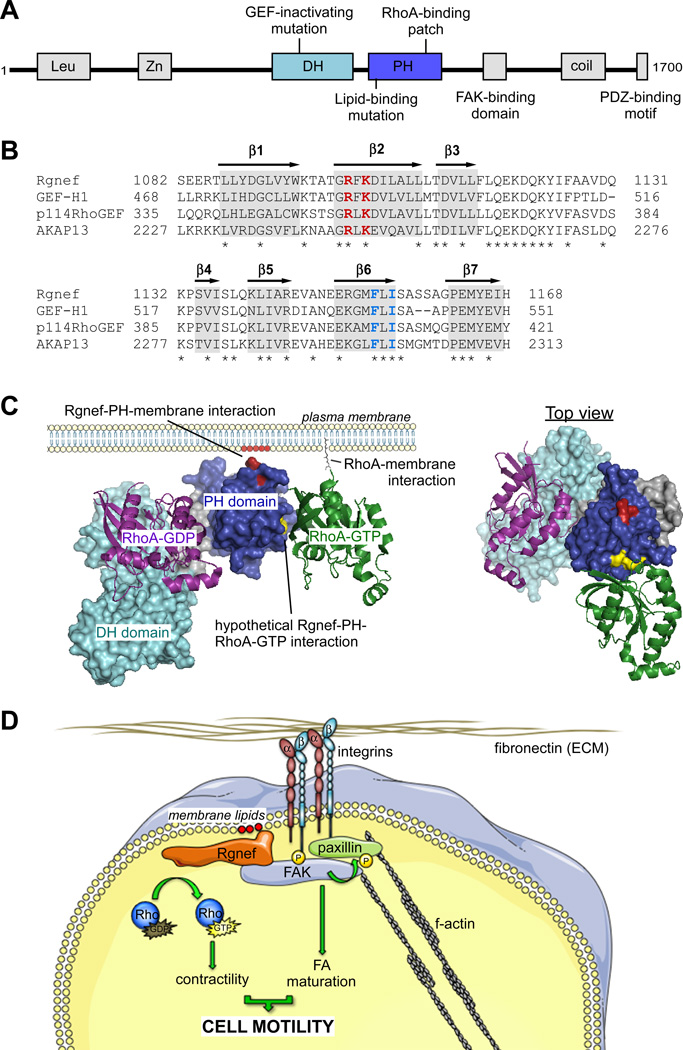
Fig. (1). Rgnef protein domains and structure.
(A) Mouse Rgnef protein schematic. Shown are the leucine-rich domain (Leu), zinc-finger motif (Zn), tandem Dbl-homology (DH) and pleckstrin-homology (PH) domains, FAK-binding domain (1292–1301), coiled-coil domain (coil), and PDZ-binding motif. Also shown are the locations of the GEF-inactivating mutation (Y1003A), lipid-binding mutation (R1098A/K1100A), and RhoA-GTP binding residues (A1151/A1153). (B) PH domain alignment of Lbc RhoGEF subfamily members. Highlighted in gray are putative locations of beta-strands (b1–b7), asterisks indicate identical residues. In red is the location of residues necessary for efficient PI lipid binding in Rgnef. In blue are residues necessary for binding to activated RhoA across all Lbc subfamily GEFs. (C) Left, theoretical structure of the Rgnef DH-PH domain at the plasma membrane. Rgnef binds to PI lipids (red) at the plasma membrane through conserved residues in the PH domain (residues in red, PH domain in blue). Rgnef also potentially binds to RhoA-GTP (green) at the plasma membrane through conserved hydrophobic residues (yellow) in the PH domain. These factors potentially localize and orient Rgnef for its GEF activity towards RhoA-GDP (purple) through the DH domain (cyan). Right, top down view of Rgnef in complex with RhoA-GTP and RhoA-GDP. Theoretical Rgnef DH-PH model created in Swiss-Model. RhoA-GDP crystal structure from PDB 1X86. RhoA-GTP crystal structure from PDB 3KZ1. Theoretical Rgnef DH-PH model created in Swiss-Model based on PDB 3KZ1 [97] (D) Simplified model of Rgnef function downsteam of integrin signaling. Cell binding ECM leads to integrin clustering and activation at the membrane, generating increased phoshatidylinositol lipids at adhesion sites. Rgnef PH domain associates with concentrated membrane lipids and facilitates FAK localization at nascent adhesions. FAK activation promotes FA maturation and Rgnef RhoA-GEF catalytic activity promotes actomyosin contractility, both required for proper cell motility.