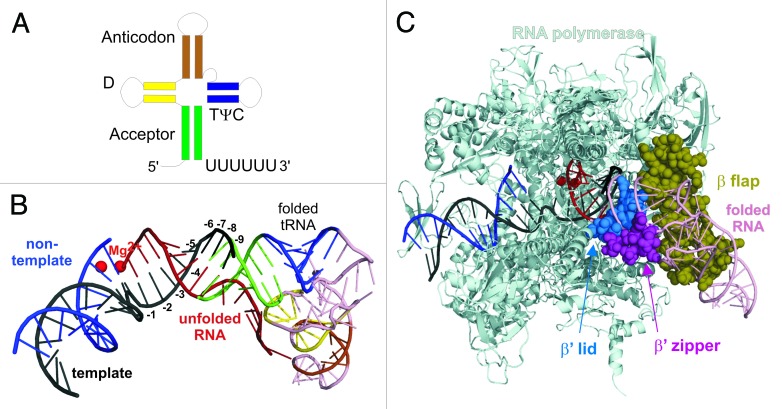
Figure 1. RNA secondary structure-dependent termination of transcription. (A) Scheme of the tRNA secondary structure. Note that both acceptor stem and TψC stem-loop can serve as termination secondary structures depending on the extent of pol III backtracking on the oligoT signal (see text). (B) Nucleic acids scaffold in the elongation complex (pdbid: 2PPB).14 Mg2+ ions of the active center are shown as red spheres. Template and non-template DNA are black and dark blue, respectively; RNA is red. A pol III transcript, represented here by tRNA molecule (pdbid: 1EHZ),15 folded at the distance sufficient for termination, is color coded as in panel A (with loops in pink). Note the interference of 5′ end nucleotides of the tRNA with the template DNA bases at positions 8th and 9th in the RNA–DNA hybrid. The real orientation of the folded tRNA relative to the hybrid may be different. More base pairs of the hybrid could be melted by the folded tRNA due to the collision of tRNA with protein domains, which could exert a pulling force on the hybrid. (C) Interference of the folded tRNA (pink) termination structure with domains of RNAP (cyan ribbon). β flap, β’ zipper, and β’ lid are shown as khaki, magenta and blue spheres, respectively. Relative orientation of tRNA as in panel B. The real orientation may differ, leading to collisions with even more domains of RNAP.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.