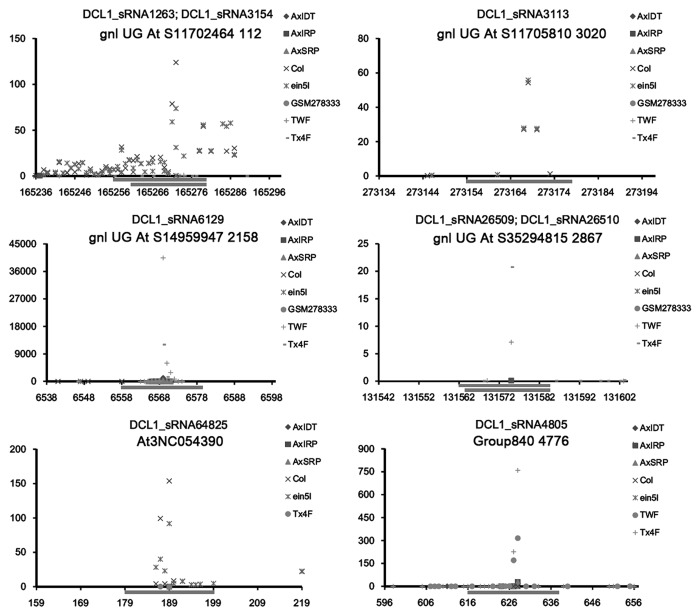
Figure 4. Degradome sequencing data-based identification of the lncRNAs (long non-coding RNAs) targeted by the DCL1 (Dicer-like 1)-dependent, AGO1 (Argonaute 1)-enriched sRNAs (small RNAs) in Arabidopsis. Examples of identified targets are shown. For each figure panel, the lncRNA ID and the corresponding sRNA(s) are listed on the top. The y axis measures the intensity (in RPM, reads per million) of the degradome signals, and the x axis indicates the position(s) of the cleavage signal(s) on the lncRNA. The binding site of the sRNA regulator on the lncRNA was denoted by a gray horizontal line. The figure keys for the signatures from different degradome sequencing libraries are shown on the right.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.