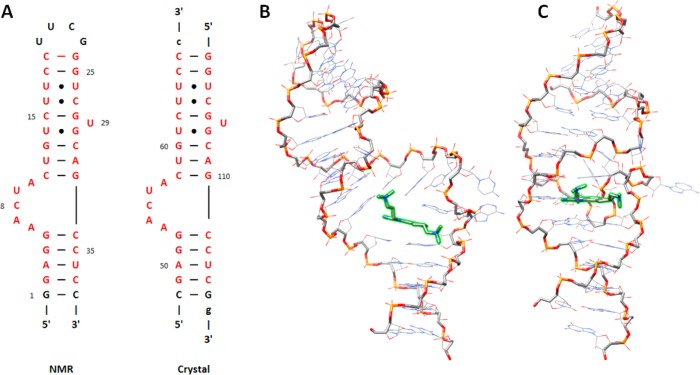
Figure 2.
Despite similar sequences, the reported conformations of the inhibitor-bound HCV IRES domain IIa determined by NMR analysis and X-ray crystallography differ. (A) Secondary structure diagrams of the domain IIa constructs used in the NMR12 and crystallography13 studies. The hairpin sequence from the NMR study was used for all of the simulations in this study. The residues colored in red show the portions of the RNA that are identical in the two published structures. (B, C) Representative models depicting the global structures of the NMR ensemble and the crystal structure, respectively. In each structure, the RNA backbone is emphasized with heavier width, and the inhibitor is highlighted in green. The structural orientations were chosen to emphasize the global differences in the binding conformations.