Figure 2.
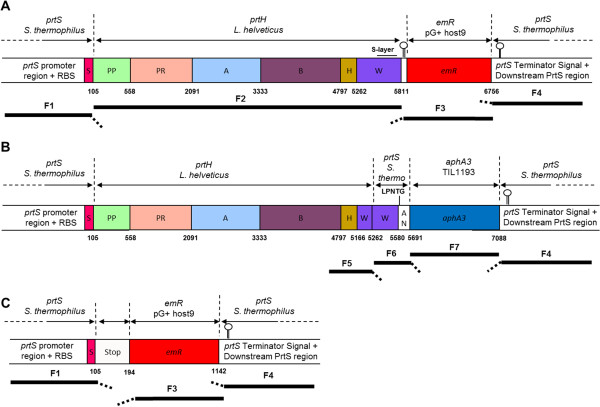
Schematic representation of the fragments used for mutant constructions and resulting mutant loci. Mutant constructions were performed by fusing different fragments from prtS, prtH, emR and aphA3 genes named F1 to F7 (black bars) to produce a PrtH fusion protein. Fragments tails (dashed thick bars) are homologous to the beginning of adjacent fragment in the construction. Numbers in bold characters refer to the last nucleotide of a domain. Nucleotides are numbered starting from the adenosine nucleotide of the start codon of the ORF. A) PrtH+ construction was inserted in S. thermophilus LMD-9 to express L. helveticus CNRZ32 CIRM-BIA 103 CEP (PrtH), instead of its own CEP (PrtS). In prtS chromosome locus, prtS promoter, prtS RBS and prtS S domain were fused to prtH gene composed of PP (propeptide domain), PR (catalytic domain), A (A domain), B (B domain), H (helix domain), W (cell-wall spacer domain) with a 303 nucleotides (nt) S-layer attachment domain (small bar) and a downstream region with the prtH terminator signal. An erythromycin resistance gene (emR) from pG+host9 [48] was inserted between prtH and the downstream region of prtS. B) PrtH+WANS construction replaced the last 450 nt, containing S-layer attachment of prtH W domain and emR gene from PrtH+ mutant, by prtS W and AN domains and aphA3 (kanamycin resistance gene) from S. thermophilus TIL1193 strain [18]. The secreted fusion PrtH+WANS proteinase has been designed to be covalently anchored to the host peptidoglycan thanks to the PrtS LPNTG motif. The PrtS W domain was expecting to bring the fused proteinase above the cell-wall of S. thermophilus. C) PrtS- sequence was obtained from a clone selected among PrtH+ potential mutants where the F2 fragment (prtH ORF) was not inserted. A 89 nt sequence composed by the two F2 primers (with 2 mutations) replaced F2 fragment in the construction.
