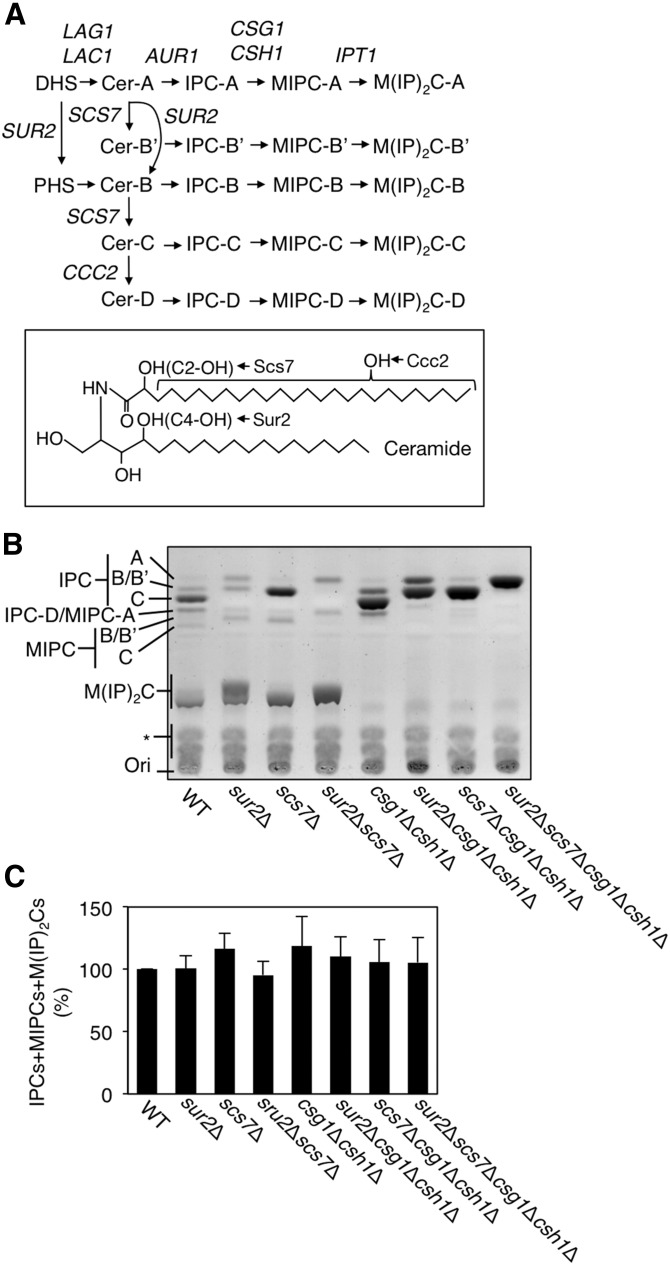
Fig. 1.
A: Comparisons of sphingolipid composition in a series of yeast mutants. Shown are the pathways for de novo sphingolipid biosynthesis and the genes involved at each step. Because of the different hydroxylation states of ceramide (Cer-A, B′, B, C, and D), there are five species each for IPC, MIPC, and M(IP)2C. B: Total lipids were extracted from 3.5 OD600 units of KA31-1A (WT), SUY12 (sur2Δ), SUY16 (scs7Δ), SUY359 (sur2 Δscs7Δ), SUY65 (csg1Δcsh1Δ), SUY401 (sur2Δcsg1Δcsh1Δ), SUY54 (scs7Δcsg1Δcsh1Δ), and SUY434 (sur2Δscs7Δcsg1Δcsh1Δ) cells and treated with mild alkaline solution to hydrolyze glycerophospholipids. The lipids were separated on a TLC plate and complex sphingolipids were visualized using 10% copper sulfate-8% orthophosphoric acid reagent. C: The expression levels of total complex sphingolipids (IPCs + MIPCs + M(IP)2Cs) were quantified with ImageJ software. The quantitative data are expressed as mean values with SDs from more than three independent experiments.