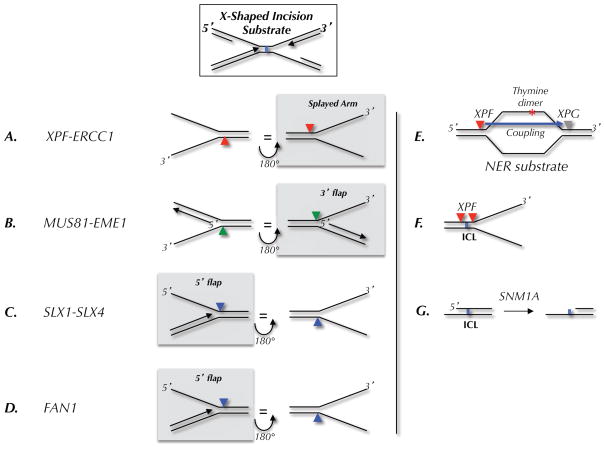
Figure 2. Model incision substrates for endonucleases.
(A–D) Preferred substrates, as determined in biochemical assays, for the indicated endonucleases. The arrowheads indicate the location of incision. Each substrate is shown in two orientations related by a 180 degree rotation. The left column shows where the nuclease would cut on the 5′ side of a converged fork; the right column shows incisions on the 3′ side. In each column, the nucleases that are likely to cut the ICL-associated X-shaped structure shown at the top are highlighted by a grey box. (E) Classic nucleotide excision repair (NER) substrate and locations of XPF and XPG incisions. (F) Location of incisions when XPF is presented with an ICL-containing fork-shaped structure. (G) Action of SNM1A on ICL-containing DNA.