Figure 3.
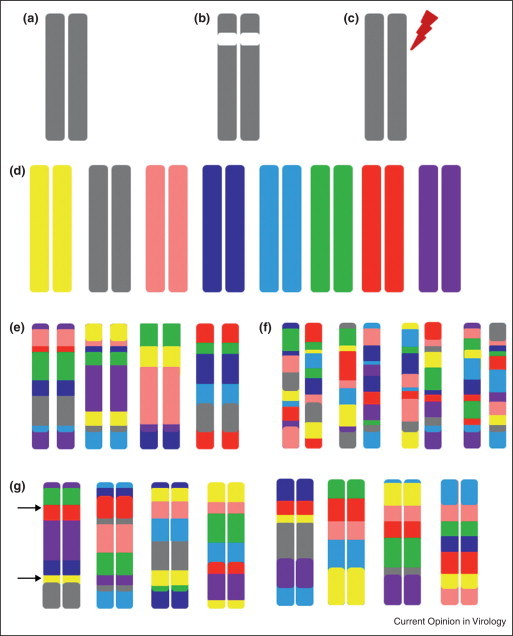
Platforms for Systems genetics discovery and validation. Traditionally, classical inbred strains such as C57BL/6J (a) have been used for systems biology approaches. These classical systems have utilized (b) gene knockouts or (c) the introduction of functional changing mutations as perturbation/validation systems. The Collaborative Cross (CC) and DO (DO) populations were derived from a set of eight genetically diverse founders whose genomes are represented by the following colors (d): A/J (yellow), C57BL/6J (gray), 129s1/SvImJ (pink), NOD/ShiLtJ (dk. blue), NZO/HILtJ (lt. blue), CAST/EiJ (green), PWK/PhJ (red), and WSB/EiJ (purple). CC lines (e) have inbred genomes that are mosaics of these eight founders (with the founder contributions keeping the color coding of D). CC lines have well-characterized genomes and being inbred are an infinitely reproducible population. Similarly (f) the Diversity Outbred (DO) is a completely outbred population of animals derived from the same eight founder strains. While this population is not reproducible, the genetic architecture of the population can be reproduced. In these ways, both the CC and DO facilitate systems genetics approaches. The CC and DO, by virtue of the large number of unique genomes, can be used (f) to create a variety of validation crosses, or sets of lines with unique genetic combinations for further mechanistic study of polymorphisms of interest. Here, a panel of CC lines is being used to contrast the PWK/PhJ (red) and 129S1/SvImJ (pink) alleles at Locus 1, while simultaneously being used to contrast A/J (yellow) and WSB/EiJ (purple) alleles at Locus 2.
