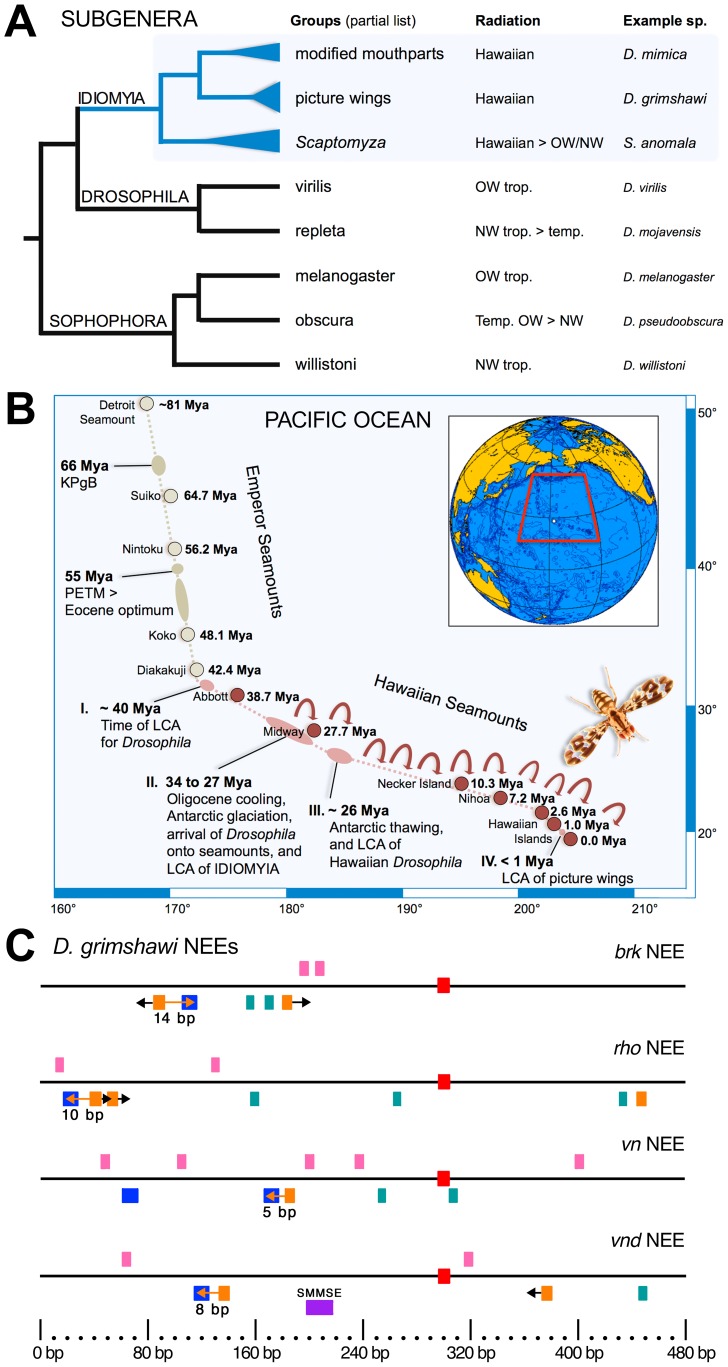
Figure 1. The Neurogenic Ectoderm Enhancers from the Hawaiian species D. grimshawi.
(A) Shown is a phylogenetic tree showing the relationship of the Hawaiian Drosophila, which comprise subgenus IDIOMYIA along with the so-called Scaptomyza [16]. This subgenus gave rise to multiple clades (highlighted in blue) corresponding to multiple adaptive radiations associated with the last ∼30 My history of island formation. Embryonic enhancers from two of these clades, the modified mouthparts group and the picture wing group, are analyzed in this study. (B) Shown is a map of the Hawaiian Seamounts with relevant events in the evolutionary lineage leading to the Hawaiian Drosophila groups indicated by the ages of the various islands. The picture wing group (inset shows an example D. grimshawi adult fly) is one of the most recent radiations and is closely associated with the newest and easternmost front of the seamounts, i.e., the Hawaiian islands. (C) The graphs indicate the site architecture of the four genus-canonical Neurogenic Ectoderm Enhancers (NEEs), which we identified in the D. grimshawi genome. The NEEs are a class of early embryonic enhancers downstream of the Dorsal morphogen gradient [4], [5], [19]. The search for NEEs in the D. grimshawi genome was conducted by searching for linked sites for the Twist:Daughterless bHLH heterodimer (Twi:Da) and the rel homology domain-containing TF Dorsal (5′-CACATGT 0–41 bp GGAAABYCC), plus a nearby Su(H) site binding site located up to +/−300 bp away (see Material & Methods). Three horizontal tracks indicate sequences matching different TF binding motifs (tracks are separated to avoid overlap and for ease of visualization when viewed in black & white print or with color-blindness). Above line: pink boxes = Snail (5′-CARRTG) [57]. On line: red boxes = Su(H) (5′-YGTGRGAA). A single Su(H) site is present in each enhancer and is used to anchor each sequence at basepair position 300 bp [the Su(H) motif matches the top strand for all except in the vn NEE]. Below line: orange boxes = Twist:Daughterless (Twi:Da, 5′-CACATGT); blue boxes = Dorsal (5′-VGGAAABYCCV); blue and orange boxes connected by arrow = linked Twi:Da–Dorsal sites with text indicating spacer length; and purple boxes = Shnurri:Mad:Medea Silencer Element (SMMSE). The Schnurri/Mad/Medea Silencer Element (SMMSE), which functions to constrain the dorsal border of activity for the NEE at vnd [19], matches the D. melanogaster/D. grimshawi consensus 5′-MYGGCGWCACACTGTCTGS and is highlighted in purple.