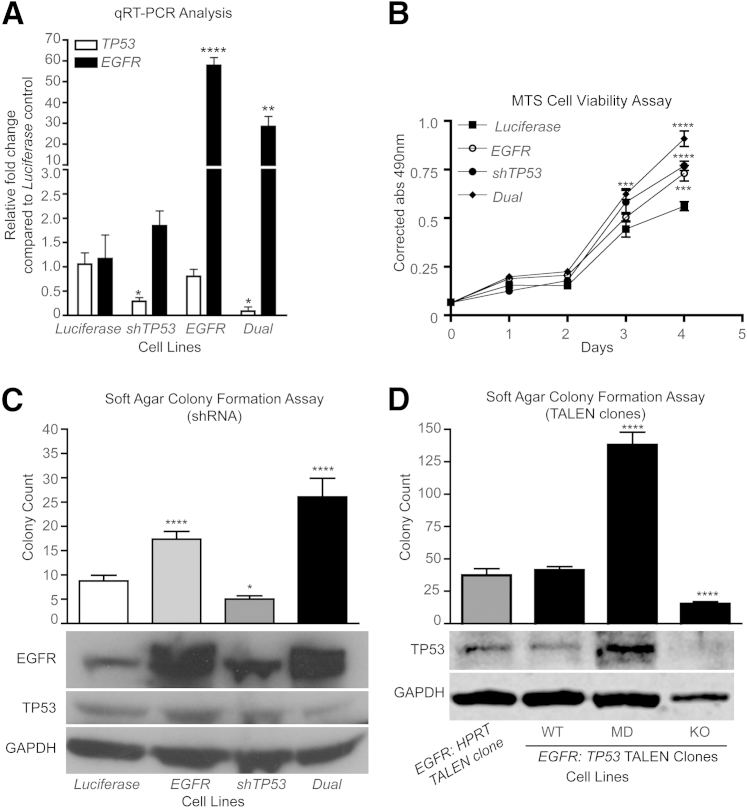
Figure 2.
Combined EGFR overexpression and reduced TP53 expression in iHSC1λ cells increase proliferation and anchorage-independent growth. A: RT-qPCR for EGFR and TP53 expression in each iHSC1λ cell line targeted with EGFR overexpression (EGFR), a TP53 shRNA (shTP53), or both (dual). A luciferase cDNA (Luciferase) served as a control. Statistical analysis performed with Student's t-test relative to Luciferase control. B: Graph depicts results from an MTS cell viability assay during the course of 4 days for each of the four cell lines. Data are a representative of three independent experiments. Statistics were performed using an unpaired Student's t-test. C: Bar graph depicts results from a soft agar colony formation assay. Experiments were performed in triplicate (n = 12 measurements for each experiment). Statistics were performed using an unpaired Student's t-test relative to a Luciferase control. Western blot analysis depicts EGFR, TP53, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each of the four cell lines. D: Bar graph depicts results from a soft agar colony formation assay. Experiments were performed in triplicate (n = 12 measurements for each experiment). Statistics were performed using an unpaired Student's t-test relative to an EGFR cell line targeted with HPRT TALENs (EGFR). EGFR:TP53 TALEN clones listed include clones in which the TP53 locus was not modified (WT), multiple mutations in different alleles are present (MD), and homozygous mutations that knocked out TP53 expression (KO). Statistics were performed using an unpaired Student's t-test relative to EGFR:HPRT control. Western blot analysis depicts TP53 and GAPDH expression in the EGFR-overexpressing cell lines. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005, and ∗∗∗∗P < 0.0001.