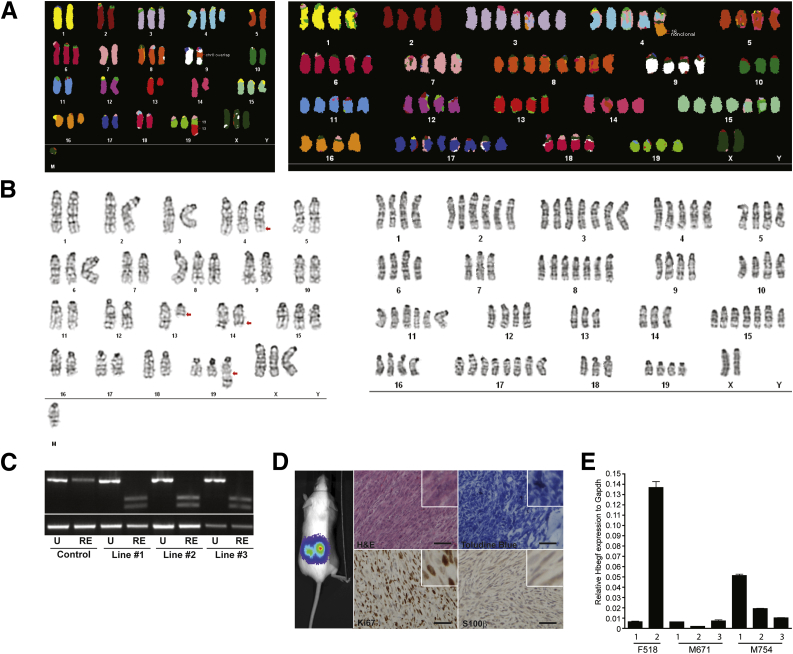
Figure 6.
Characterization of grade 3 PNST-derived cell lines. A: SKY of grade 3 PNST-derived cell lines. Two clones from a single tumor are depicted. The clone on the left possesses translocations (chromosomes 9 and 19), duplications (chromosomes 3, 4, 6, 8, 15, 16, 19, and X), and deletions (chromosome 14). On the right, a clone possesses large whole chromosomal amplifications (chromosomes 1 to 19) and a translocation on chromosome 4. B: G-banding analysis of grade 3 PNST-derived cell lines. Both clones are from the same tumor in A. The clone on the left possesses a few deletions, amplifications, and translocations (arrows). The clone on the right possesses numerous whole chromosomal amplifications. C: Agarose gel electrophoresis of PCR products from a Trp53 loss-of-heterozygosity experiment. D: A band at 600 bp represents the WT Trp53 allele, whereas the double smaller bands represent the digested Trp53R270H allele. Luciferase live imaging of cell lines injected into SCID/BIEGE mice demonstrating their capacity for tumor formation. Histological analysis of tumors for H&E staining, toluidine blue staining, and IHC for Ki-67 and S100β indicates the cell lines form grade 3 PNSTs. E: Bar graph depicts RT-qPCR results for Hbegf mRNA expression relative to Gapdh on grade 3 PNST-derived cell lines from three independent tumors. Original magnification, ×400 (D). Scale bar = 50 μm (D). RE, restriction enzyme digested; U, uncut.