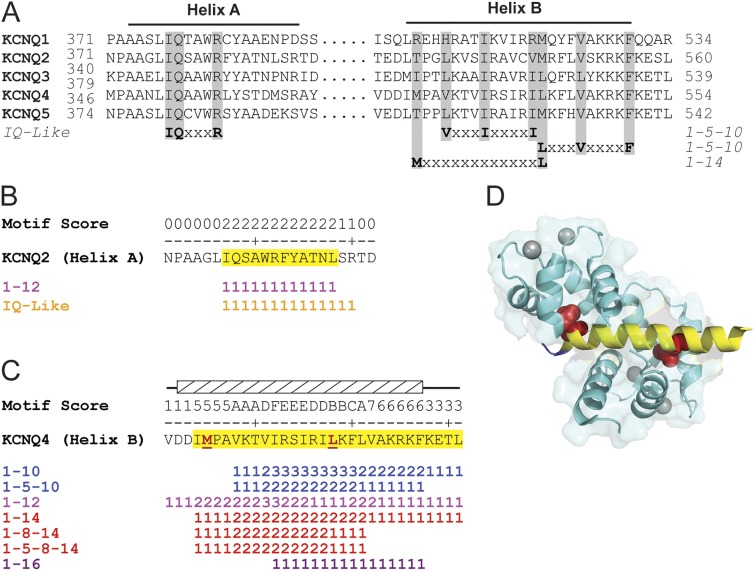
Figure 1.
Meta-analysis of KCNQ CaM-binding regions identifies multiple canonical motifs that map to the crystallized structure. (A) Sequence alignment of helices A and B. Gray shading demarcates key residues that define the previously identified CaM-binding motifs. “Wiggle plot” depiction of all of the canonical CaM-binding motifs within (B) KCNQ2 helix A and (C) KCNQ4 helix B. Yellow highlighted residues denote the meta-analysis CaM-binding domain predictions. Hatched box and red underlined residues denote target peptide and anchor residues shown in D. Motif score equals the number of times an amino acid is found in a unique canonical CaM-binding motif. (D) Ribbon diagram of CaM–KCNQ4 helix B (PDB accession no. 4GOW) with the meta-analysis prediction (yellow) mapped onto the target peptide (blue). Red space fill, anchor residues; cyan with gray calcium ions, CaM.