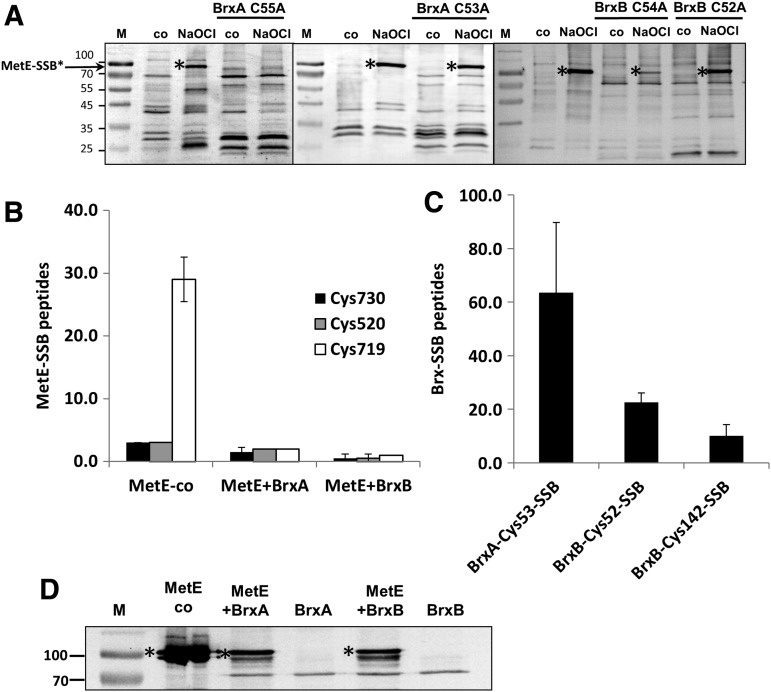
FIG. 5.
De-bacillithiolation of MetE-SSB by BrxA and BrxB. (A) S-bacillithiolated MetE (MetE-SSB) was generated in vivo by the treatment of cells with NaOCl for 10 min. Cell extracts were prepared, and the presence of MetE-SSB (*) was monitored by immunoblotting with anti-BSH antibodies as previously described (4, 5). The prominent ∼90 kDa band in NaOCl-treated cells corresponds to MetE-SSB, and other bands are other S-bacillithiolated proteins as previously documented by MS (4, 5). Incubation of purified BrxA C55A (∼15 μM) for 30 min with crude cell extracts eliminated the MetE-SSB signal, and BrxB C54A greatly reduced the signal intensity. In contrast, BrxA C53A and BrxB C52A did not detectably de-bacillithiolate MetE-SSB. The left lane in each panel contains size markers. (B–D) To test the ability of native BrxA and BrxB proteins to de-bacillithiolate MetE-SSB, and to determine which site(s) of modification are affected by Brx proteins, we prepared MetE-SSB in vitro by the incubation of MetE with oxidized BSH (BSSB). MetE-SSB modification was monitored by MS and was found to be predominantly on Cys719 with lesser amounts of modification on Cys520 and Cys730. Incubation with 15 μM BrxA and BrxB (after reduction with NaBH4) led to nearly complete de-bacillithiolation of MetE-SSB as revealed by the quantitation of spectral counts in the MS analysis (B) and by BSH immunoblots (D). Concomitant with MetE-SSB de-bacillithiolation, both BrxA and BrxB became S-bacillithiolated at their active site Cys residues with a minor amount of S-bacillithiolation also detected on BrxB Cys142 (C). BSSB, oxidized bacillithiol disulfide; MS, mass spectrometry.