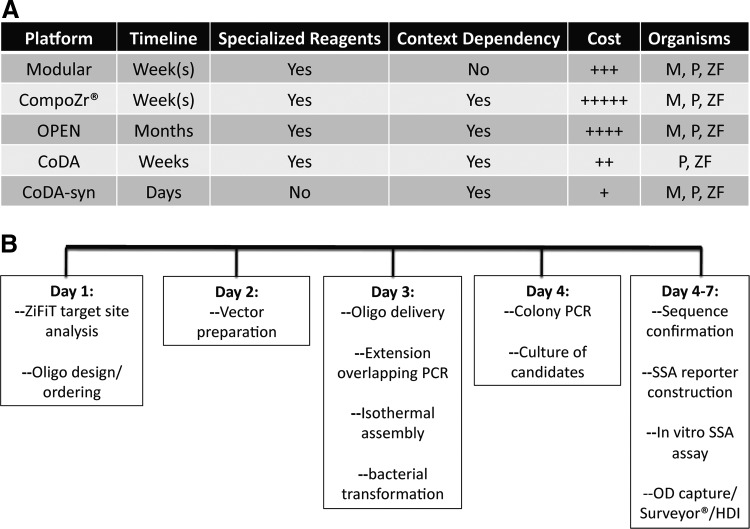
FIG. 7.
CoDA-syn procedure comparison to alternate ZFN generation methods. (CoDA-syn procedure comparison to alternate ZFN generation methods. (A) Multiple platforms exist for generating gene-specific ZFNs. A generalized time line, the necessity for acquiring/purchasing specific starting materials, whether context dependency is inherent to the generation, a generalized cost analysis, and ability to modify genes in multiple organisms are provided. (B) CoDA-syn assembly schema. The in silico gene analysis and oligonucleotide design and ordering can be accomplished in 1 day. Before oligo delivery the expression plasmid vector and middle fragments can be prepared by restriction digest and gel purification and archived. After oligo delivery, they are resuspended, pooled, and subjected to overlapping extension PCR that yields a 300-bp product. The purified vector and PCR fragments are added to the isothermal DNA assembly mixture and incubated at 50°C for 30 min and then transformed into Epi300 bacteria. On day 4, 10 colonies are chosen for colony PCR followed by overnight culture. Plasmids are sequenced while the SSA reporter is generated. Once full sequence confirmation is received the in vitro analysis of synthetic ZFN candidates can be performed in the cell type of choice using the SSA, OD, Surveyor, and Homology Directed Integration (HDI) assays. M, mammalian; P, plant; ZF, zebrafish.