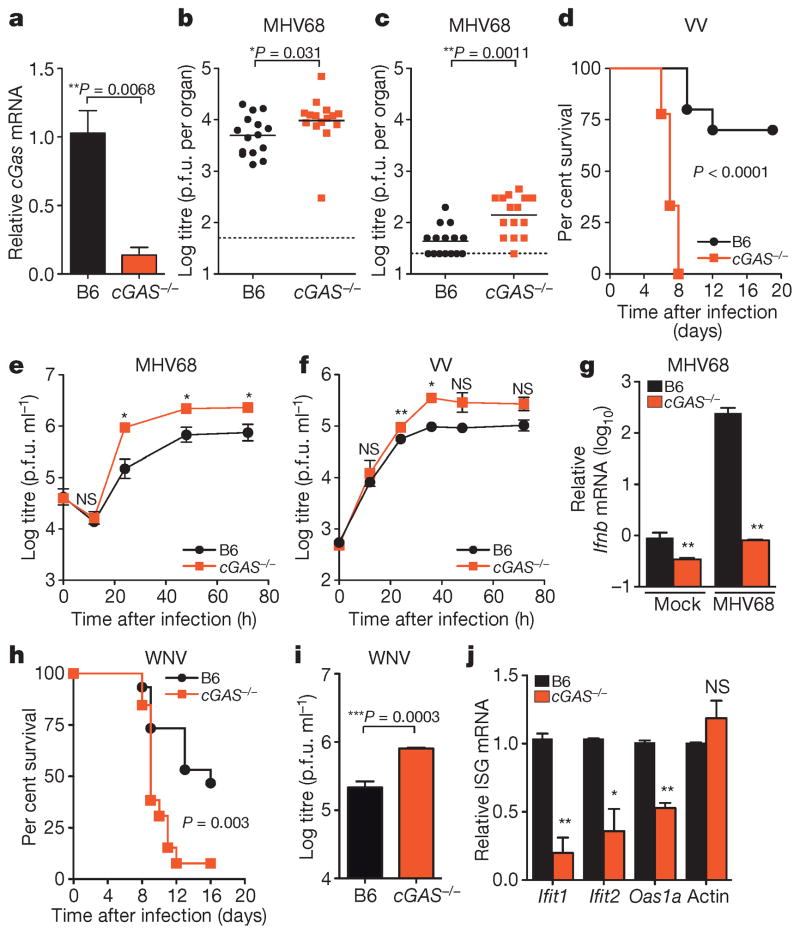
Figure 4. Requirement for cGAS in controlling viral infection in vivo.
a, Quantitative PCR with reverse transcription (qRT–PCR) of relative cGas expression in spleens of wild-type (B6) and cGas−/− mice. n = 3 mice per group. Error bars represent s.e.m. Statistical significance was determined by t-test. b, c, Viral titres in spleen (b) and lungs (c) of wild-type and cGas−/− mice after infection with 106 plaque-forming units (p.f.u.) of MHV68, n = 15 mice per group. Statistical significance was determined by Mann–Whitney test. d, Survival curves of wild-type and cGas−/− mice infected with 8000 p.f.u. of VV, n = 10 (wild type), n = 9 (cGas−/−). Statistical significance was determined by log rank test. e, f, i, Viral titres from BMMO after infection with 10 multiplicity of infection (m.o.i.) MHV68 (e), 0.1 m.o.i. VV (f) or 0.1 m.o.i. WNV (i). Data represent three (e, f) or four (i) independent experiments performed in triplicate. Error bars represent s.e.m. Statistical significance was determined by t-test. g, qRT–PCR of relative Ifnb expression in BMMO infected with MHV68 for 6 h. Data represent means of three experiments performed in triplicate. Error bars represent s.d. Statistical significance was determined by t-test. h, Survival curves of wild-type and cGas−/− mice infected with 100 p.f.u. of WNV, n = 10 (wild type), n = 9 (cGas−/−). j, qRT–PCR of baseline gene expression (Ifnb, Ifit1, Ifit2, Oas1a, actin) in wild-type and cGas−/− BMMO. Data represent means of two experiments performed in triplicate. Error bars represent s.d. Statistical significance was determined by t-test. In all panels, *P <0.05, **P <0.01, ***P <0.001; NS, not significant.