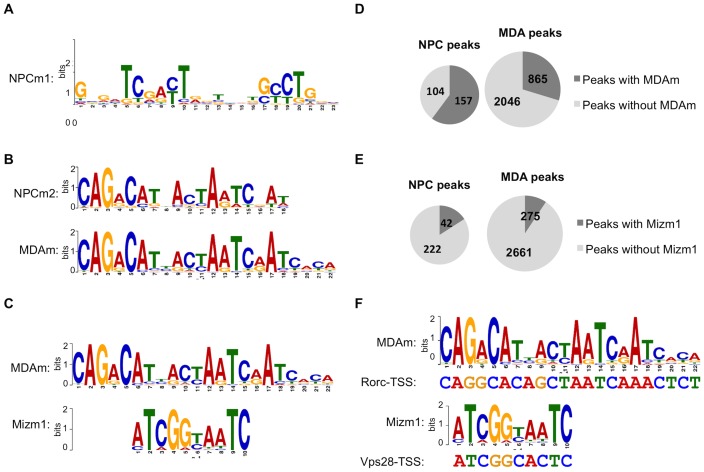
Figure 8. Motifs enriched in Miz-1 ChIP-seq peaks match Mizm1.
(A) The top-scoring peak in NPCs, as reported by Wolf et al., has little similarity to Mizm1. (B) Tomtom alignment of the top-scoring motif in MDA peaks (MDAm) with the second-top scoring motif in NPC peaks (NPCm2), demonstrating that they are nearly identical to each other (p = 8.8×10−10). (C) Tomtom alignment of Mizm1 with the central portion of MDAm, showing statistically significant similarity (p = 0.009). (D) FIMO was used to identify ChIP-seq peaks containing instances of MDAm in the NPC (left) and MDA (right) Miz-1 ChIP-seq peak sets, using a cutoff of p<0.0001. (E) FIMO was used to identify ChIP-seq peaks containing instances of Mizm1 in the NPC (left) and MDA (right) Miz-1 ChIP-seq peak sets, using a cutoff of p<0.0001. (F) FIMO was used to search the ChIP-seq peaks that Wolf et al. validated by ChIP-qPCR for matches to the MDAm and Mizm1 motifs. Examples of statistically significant matches are shown (p = 1.7×10−7 for MDAm in RORC-TSS; p = 1.22×10−5 for Mizm1 in Vps28-TSS).