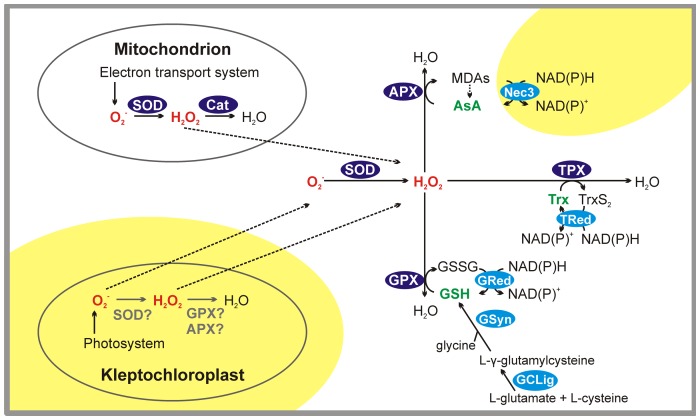
Figure 4. Hypothetical pathways for reactive oxygen species (ROS) detoxification.
Pathways evidenced in both ciliates (white background) or only in Strombidium rassoulzadegani (yellow background) are shown. In red, ROS (O2 - = superoxide radical; H2O2 = hydrogen peroxide); in dark blue, antioxidant enzymes (SOD = superoxide dismutase; Cat = catalase; APX = ascorbate peroxidase; GPX = glutathione peroxidase; TPX = thioredoxin peroxidase); in light blue, enzymes involved in metabolism of non-enzymatic antioxidants (Nec3 = bifunctional monodehydroascorbate reductase/carbonic anhydrase nectarin-3-like; TRed = thioredoxin reductase; GRed = glutathione reductase; GSyn = GSH synthetase; GCLig = Glutamate-cysteine ligase); in green, non-enzymatic antioxidants (AsA = ascorbic acid; GSH = glutathione; Trx = thioredoxin). MDAs = monodehydroascorbate; GSSG = glutathione disulfide; TrxS2 = thioredoxin disulfide; NAD(P)+ = nicotinamide adenine dinucleotide (phosphate). Based on literature [61], [63], [75].