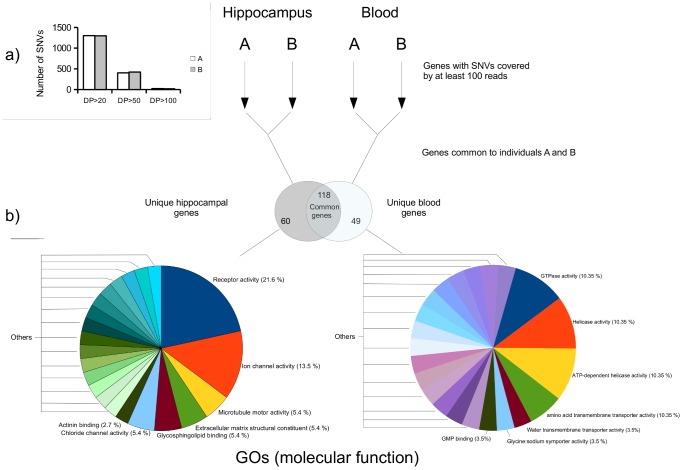
Figure 4. GO terms for genes with highly covered tissue-specific SNVs.
a) (Inset) Number of hippocampus-specific SNVs (present in hippocampus, but not in blood) for subjects A and B, taking account the number of reads covering each SNV: at least 20 (DP20), 50 or more (DP50), and 100 or more (DP100). b) Analysis of GO terms for genes with highly covered SNVs. We selected lists of genes common to subjects A and B with at least one SNV in hippocampus and blood, covered by at least 100 reads, in order to test for differences in the annotations of the GOs of genes with SNVs tissue-specific. The genes with SNVs in hippocampus and blood covered by at least 100 reads were analyzed by examining genes exclusive to the hippocampus and those exclusive to blood. The annotations for molecular function GOs overrepresented in each list of hippocampus- (left) and blood- (right) specific genes are shown. Observe that the main annotations vary greatly for each tissue. There is overrepresentation of genes with SNVs in the hippocampus with functions of receptor activity (21.6%), ion channel activity (13.5%) and microtubule motor activity (5.4%), while for genes with highly covered SNVs in blood, the main molecular function annotations are GTPase activity (10.35%), Helicase activity (10.35%), and others.