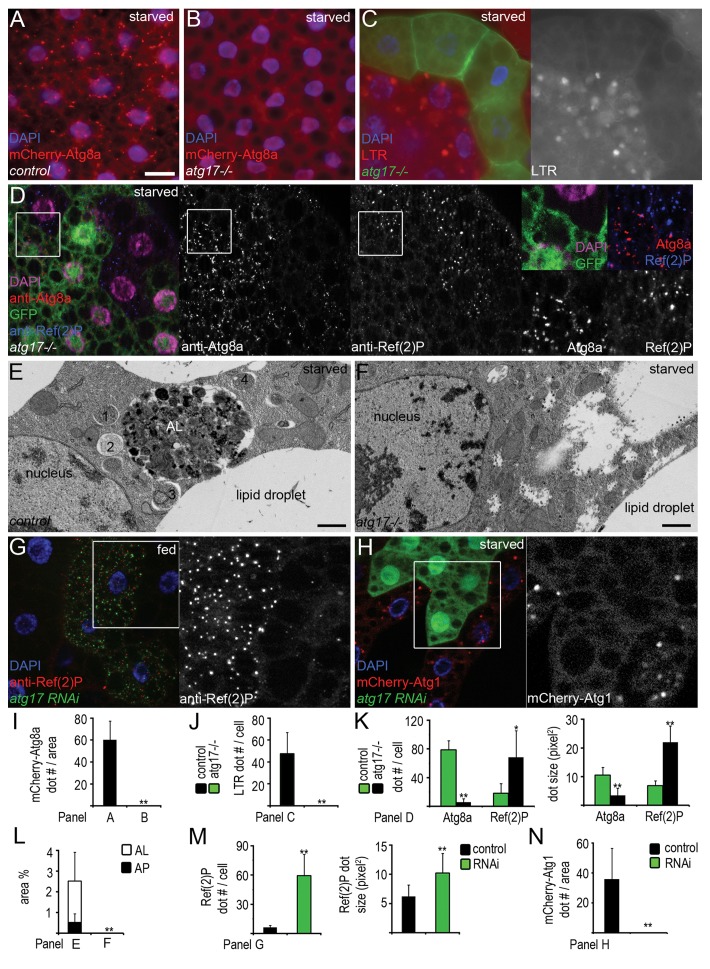
Figure 2.atg17 is required for starvation-induced and basal autophagy. (A and B) mCherry-Atg8a-positive autophagosomes and autolysosomes form in fat body cells of starved control (A) but not in atg17-null mutant larvae (B). (C) No LTR (LysoTracker Red)-positive autolysosomes form in GFP-positive atg17-null mutant cells, compared with surrounding control cells in the same fat body tissue in starved animals. (D) Endogenous Atg8a-positive autophagosomes are missing and Ref(2)P aggregates accumulate in starved atg17-null mutant cells (these are marked by the lack of GFP in this image, unlike in C), compared with neighboring control cells. (E and F) Electron microscopy shows that autophagosomes (1–4) and autolysosomes (AL) form in fat body cells of control (E) but not in atg17-null mutant larvae during starvation (F). (G) Depletion of atg17 in GFP-positive cells results in large-scale accumulation of Ref(2)P in fat bodies of well-fed larvae. (H) Depletion of atg17 prevents starvation-induced mCherry-Atg1 dot formation. (I–N) Quantification of data from (A and B) (I, n = 12/genotype), (C) (J, n = 14), (D) (K, n = 18), (E and F), showing the ratio of autophagosomes and autolysosomes relative to total cytoplasm (L, n = 4/genotype), (G) (M, n = 10), (H) (N, n = 10). Scale bar: (A–D, G, and H) 20 µm; (E and F) 1 µm. Error bars: s.d., *P < 0.05, **P < 0.01.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.