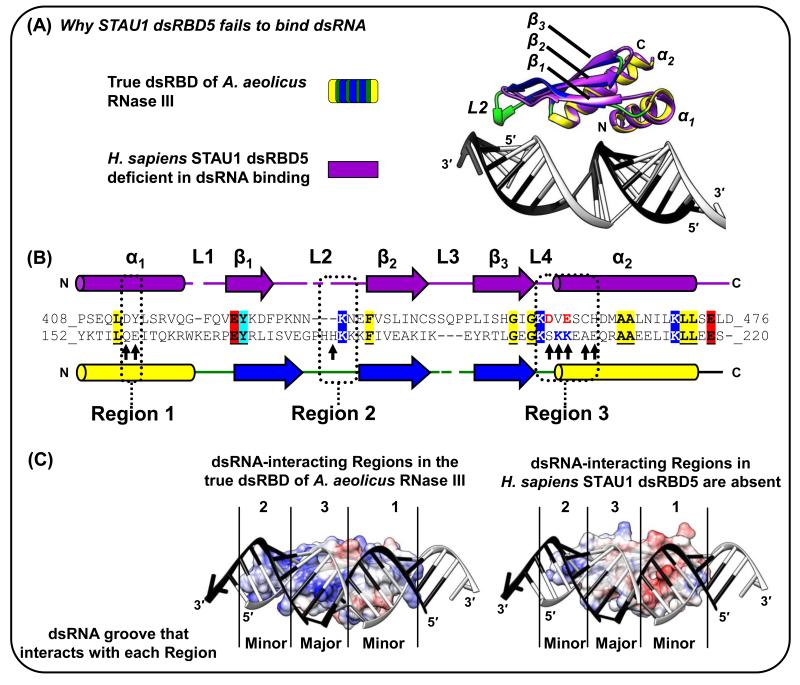
Figure 3. The STAU1 dsRBD5 type-B dsRBD can fold nearly identically to true dsRBDs but lacks all three dsRNA-interacting Regions [17].
(A) Superposition of the X-ray crystal structure of human STAU1 dsRBD5 with that of the true dsRBD of A. aeolicus RNase III (Figure 1A-D; [3, 17]). The mainchain (i.e. Cα backbone) of STAU1 dsRBD5 has root mean square deviations as low as 1.8 Å relative to true dsRBDs. (B) Structural-based alignment of dsRBDs in (A). Identical residues are shaded and underlined if they reside outside the dsRNA-interacting regions, which suggests conserved contributions to the fold rather than to dsRNA-binding directly. Arrows within Regions 1-3 are those in the A. aeolicus RNase III dsRBD 3 that match the consensus dsRBD. Notice especially the STAU1 dsRBD5 negatively charged D and E residues positioned in Region 3 near where dsRNA-interacting residues KK exist in true dsRBDs. (C) Electrostatic potential surfaces, generated using the Chimera program, of (left) A. aeolicus RNase III dsRBD (Figure 1A-D; [3]), or (right) STAU1 dsRBD5 [17] superimposed onto the A. aeolicus RNase III dsRBD. Blue, red or white signify, respectively, positively, negatively or neutral charged surfaces. dsRNA in the A. aeolicus RNase III dsRBD structure is shown for both dsRBDs to illustrate for STAU1 dsRBD5 the lack of charged residues in Region 3 and the shortened Region 2, which partially explain its inability to bind dsRNA.