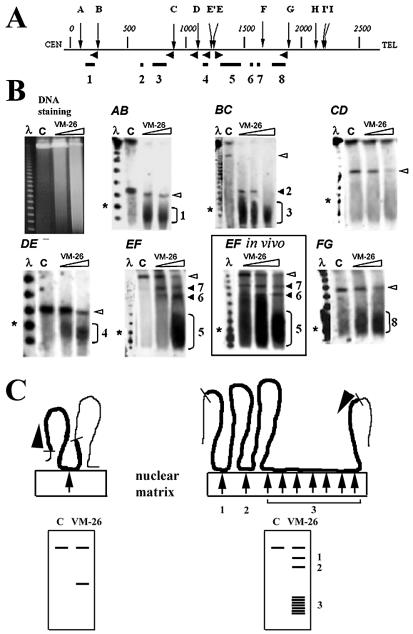
Figure 1.
Mapping DNA loops and attachment regions in the human dystrophin gene by DNA cleavage mediated by nuclear matrix-bound topoisomerase II. (A) Map of the gene with distances in kb and SfiI sites indicated by letters. Horizontal arrowheads indicate positions and directions of probes used to detect the six SfiI fragments AB, BC, CD, DE, EF and FG on PFGE gels. Loop attachment regions deduced from the truncation positions are shown on the map by the horizontal lines (1–8). (B) Separation of the products of topoisomerase II-mediated DNA cleavage by PFGE (left) and detection of SfiI fragments by hybridization. Left lanes show λ DNA oligomers, asterisks the 100 kb marker, and lanes C are DNA from control samples not treated with VM-26. Full-length SfiI fragments are indicated by open arrowheads and fragments truncated due to topoisomerase II cleavage by the numbers 1–8. The boxed insert (EF in vivo) shows cleavage of fragment EF by topoisomerase II in living cells. (C) Schematic depiction of the excision of DNA loops by nuclear matrix-bound topoisomerase II. SfiI fragments are shown in bold, topoisomerase II cleavage sites by arrows, and hybridization probes by arrowheads. The resulting DNA fragments are separated by PFGE and truncated fragments are detected by hybridization (below). A longer attachment region (right) could cause a heterogeneous cleavage pattern such as that causing the wide hybridizing band in panel EF.