Figure 4.
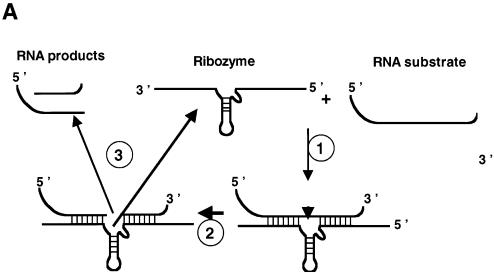
Enhancement of ribozyme cleavage by FMRP. (A) Assay schematic. RNA substrate and hammerhead ribozyme, both 32P-labelled, were generated by in vitro transcription and purified by PAGE. Cleavage of the 32P-labelled RNA substrate by the hammerhead ribozyme first necessitates hybridization of the ribozyme to the substrate (step 1). After substrate cleavage (step 2, see arrow head), the RNA products must be released to allow recycling of the ribozyme (step 3). At the end of the reaction, protein is removed by phenol extraction and 32P-labelled RNAs analysed by PAGE under denaturing conditions to visualize the 32P-labelled RNA products. In the absence of a nucleic acid chaperone, annealing of the substrate to the ribozyme and release of the RNA products appear to be slow. Addition of a nucleic acid chaperone will accelerate hybridization of the substrate to the ribozyme and dissociation of the RNA products, and thus ribozyme turnover. Base pairing between the RNA substrate and ribozyme R3 are underlined on the substrate sequence. Ribozyme-mediated cleavage occurs on the 3′ side of A (space) for RNA S14, …GAUUAAGUAGUA AGAGUGUCUGCA-3′, and for RNA S20, …GAUUAAGUAGUA AGAGUGUCUGCA-3′. (B) Ribozyme-directed cleavage of RNA substrate. Percentages of ribozyme-directed RNA cleavage at 25 min were assessed by PhorphorImaging and are indicated in parentheses (see below). The vertical arrow shows the direction of electrophoresis. Ribozyme R3 (0.3 pmol) and RNA S14 (0.1 pmol) were incubated as described in Materials and Methods. 32P-labelled RNA substrate (S14) and product (ΔS14) were analysed by denaturing 8% PAGE in 0.5× TBE. Lanes 1–2, R3 and S14 at 4 or 37°C for 30 min (5% and 20%). Reactions with proteins were for 15 min at 37°C. Lanes 3–5, HIV-1 NCp7 at protein to nucleotide molar ratios of 1:20, 1:10 and 1:5, respectively (60, 73 and 75%), corresponding to a protein concentration of 0.5, 1 and 2 × 10–7 M, respectively. Lanes 6–8, NC(12–53) at molar ratios of 1:2.5, 1:1.2 and 1:0.6, respectively (25% for all ratios), corresponding to a protein concentration of 4, 8 and 16 × 10–7 M, respectively. Lanes 9–11, FMRP at protein to nucleotide molar ratios of 1:20, 1:10 and 1:5, respectively (33, 45 and 66%), corresponding to a protein concentration of 0.5, 1 and 2 × 10–7 M, respectively. Respective concentrations of the FMRP variants (lanes 12–20) were the same (see below). Lanes 12–14, FMRP ΔPPId at molar ratios of 1:20, 1:10 and 1:5, respectively (30, 35 and 39%). Lanes 15–17, FMRP ΔKHT at molar ratios of 1:20, 1:10 and 1:5, respectively (25, 28 and 32%). Lanes 18–20, FMRP ΔRGG at molar ratios of 1:20, 1:10 and 1:5, respectively (30, 40 and 57%). R3, S14 and the 5′ sequences of S14 (ΔS14) are identified on the right. Markers are on the left. Note that the RNA products rapidly accumulate in the presence of a chaperone like NCp7 (lanes 3–5) or FMRP (lanes 9–11), whereas they do so only slowly in the absence of a chaperone (lane 2). (C) Ribozyme-directed cleavage of RNA substrate S20. Ribozyme R3 (0.3 pmol) and RNA S20 (0.1 pmol) were incubated as described in Materials and Methods. 32P-labelled RNA substrate (S20) and product (ΔS20) were analysed by denaturing 8% PAGE in 0.5× TBE. Lanes 1–2, R3 and S20 at 4 or 37°C for 30 min (5 and 25%). Lanes 3–4, HIV-1 NCp7 at protein to nucleotide molar ratios of 1:10 and 1:5, respectively (15%). Lanes 5–6, NC(12–53) at molar ratios of 1:1.2 and 1:0.6, respectively (10%). Lanes 7–8, FMRP at protein to nucleotide molar ratios of 1:10 and 1:5, respectively (16%), corresponding to a protein concentration of 1 and 2 × 10–7 M, respectively. Respective concentrations of the FMRP variants (lanes 9–14) were the same (see below). Lanes 9–10, FMRP ΔPPId at molar ratios of 1:10 and 1:5, respectively (16%). Lanes 11–12, FMRP ΔKHT at molar ratios of 1:10 and 1:5, respectively (17%). Lanes 13–14, FMRP ΔRGG at molar ratios of 1:10 and 1:5, respectively (20%).