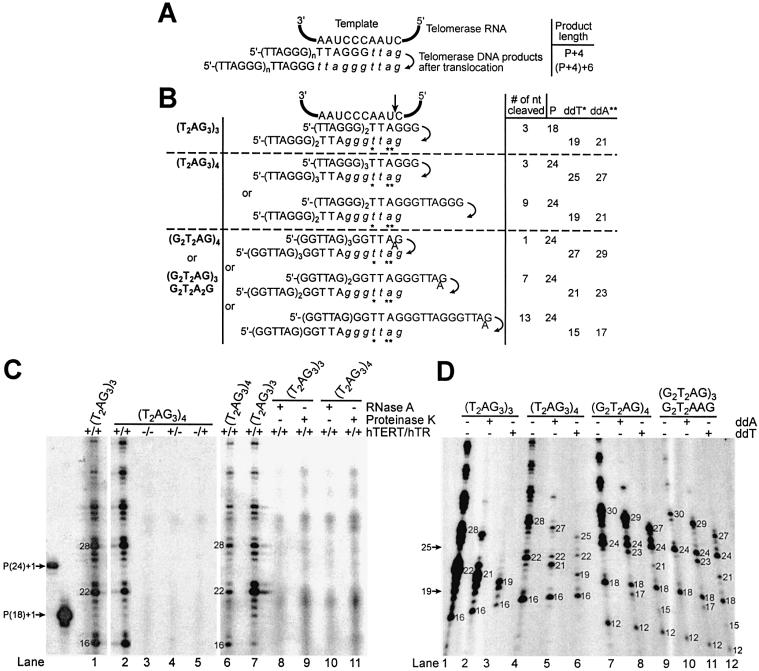
Figure 1.
Cleavage of telomeric 3′-ends. (A and B) Potential alignments of various telomeric oligonucleotides (tested in C and D) with the hTR template, before and after translocation. Compared to (G2T2AG)4, (G2T2AG)3G2T2A2G has an extra A predicted to form a mismatch with the RNA template. Italicized letters represent nucleotides added by the telomerase enzyme onto the 3′-end of the primer during the elongation step. The diagram implies that cleavage and nucleotide addition to the RNA template 5′-end precede translocation. P: primer length. ddA and ddT: dideoxyATP and dideoxyTTP. Numbers in A–D represent the lengths of the primers, elongation or chain termination products. The proposed cleavage site is shown by the vertical arrow. Asterisks indicate the positions of the first ddA or ddT incorporated during the DNA primer extension assay. (C and D) DNA primer extension assays were performed with the indicated oligonucleotides using RRL-reconstituted telomerase enzyme. The elongation products were resolved on 8% polyacrylamide-urea gels and subjected to autoradiography. The 3′-end radiolabeled 5′-biotinylated primers, (T2AG3)3 and (T2AG3)4, migrate, respectively, as 19 and 25mers [P(18)+1 and P(24)+1, respectively].