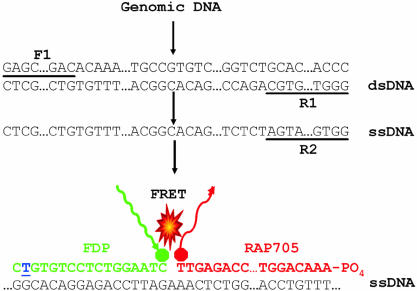
Figure 1.
Diagram showing the PCR amplification of a target sequence and detection using FRET-based DNA hybridization probes with R201C mutation. The double-stranded cDNA of the target sequence (dsDNA) was amplified with forward (F1) and reverse (R1) primers. Subsequently, a shorter single strand was amplified (ssDNA) using only one, more 5′ reverse primer (R2). DNA hybridization probes (DNA-HP) were designed in such a way that donor and acceptor probes localize in a head to tail arrangement when hybridized to ssDNA. The R201C mutation site is indicated by an underlined blue nucleotide. When the fluorescent donor probe (FDP) was excited at 470 nm, the FDP transferred its energy to the acceptor probe (RAP705) that then emitted light of 705 nm for DNA-HP. The intensity of this signal was proportional to the amount of target DNA molecule bound. Both wild-type and mutant sequences were detected by FRET in the single tube, with the mutant probe melting off at a higher temperature than the mismatched wild-type probe. Similar approaches were also taken for R201H mutation analysis. The FRET-based PNA hybridization probes with R201C and R201H mutations were also designed with similar approaches where DNA was substituted by PNA.