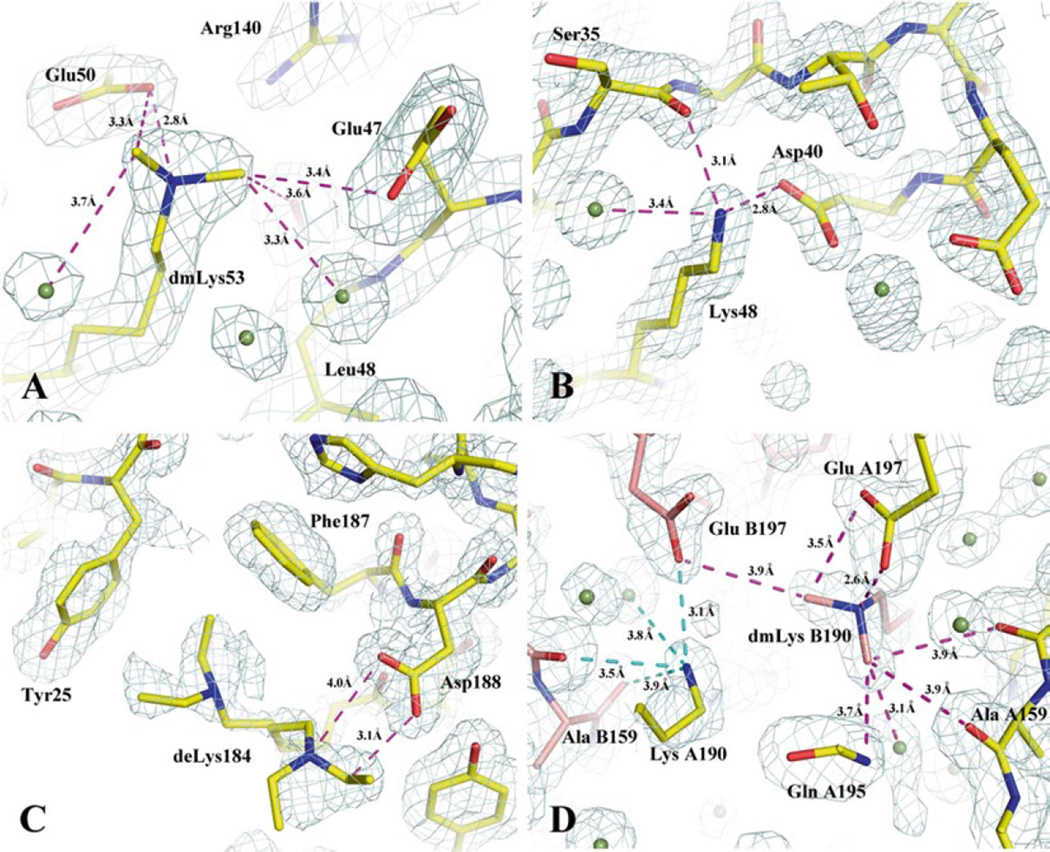
Fig. 1.
Examples of methylated, unmethylated and ethylated lysines found in alkylated protein structures and the interactions promoted by alkylated lysines. Proteins are drawn in stick format incased in navy mesh of a 2Fo–Fc map contoured at 1σ. (a) A di-methylated lysine 53 (dmLys53) interacts with carboxylates, carbonyl, and water. It is also a part of an extended hydrogen bond network. The figure was prepared based on the structure of a secreted protein from Salmonella enterica subsp. enterica serovar typhimurium str. 14028S (gi: 267994654, PDB code: 4HG1). In this case, the native protein crystals from initial screenings diffracted to about 3.5 Å. After the protein was methylated, the modified protein crystals diffracted to 1.75 Å and its structure determination was straightforward (Table 1, PDB: 4HG1). This was a project in collaboration with the Program for the Characterization of Secreted Effector Proteins (PCSEP) of the Pacific Northwest National Laboratory. (b) An unmethylated lysine (Lys48) involved in multiple interactions (salt bridge and hydrogen bonds) with other protein atoms and one water molecule in a methylated structure (PDB code: 3BED). It is believed that a lysine involved in strong intrachain interactions may prevent the residue from being methylated. The exampled structure is from the mannose/sorbose specific IIA subunit of phosphotransferase system from E. faecalis v583. (c) A diethylated lysine (deLys) in two conformations. Ethylated lysine residues tend to have multiple conformations and be at least partially disordered. In this structure from an ethylated short-chain dehydrogenase/reductase from V. parvula DSM 2008, the deLys184 forms an additional hydrogen bond to Asp188. It also makes hydrophobic contacts to Tyr25 and Phe187. The hydrophobic interaction added from ethylated lysine is believed to be its major feature. (d) In the methylated structure (PDB code: 3BED) as mentioned in (b), the two chains, (A) and (B), make a contact that is nearly twofold symmetric. Across the small interface, methylated lysine 190 from the (B) chain (dmLys B190) forms an extensive interaction to the (A) chain. Carbon atoms from the (A) and (B) chains are colored in yellow and salmon, respectively