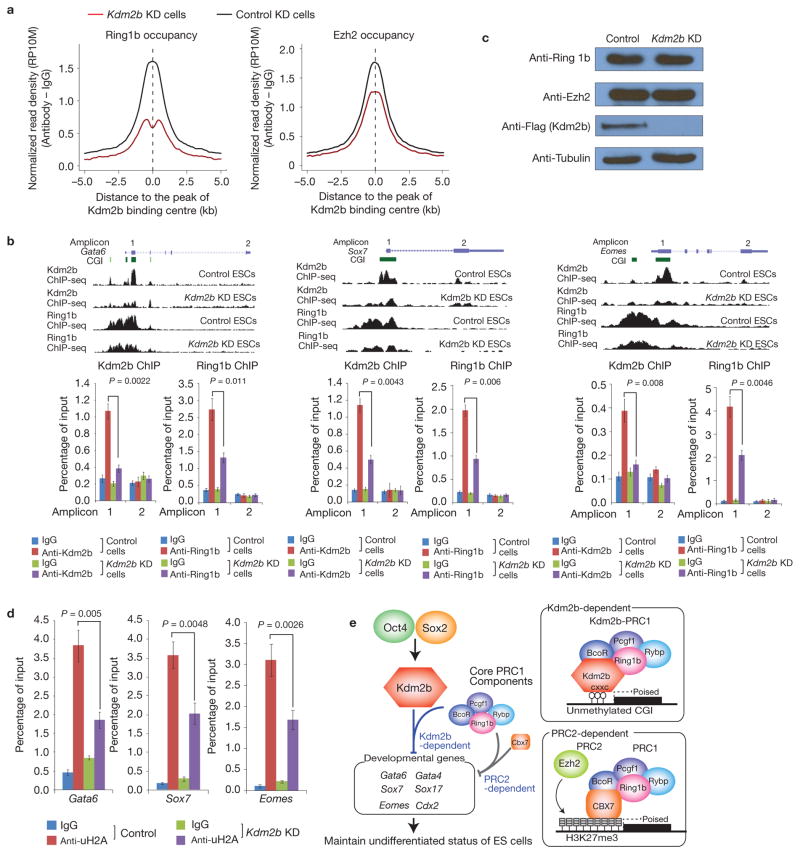
Figure 7.
Depletion of Kdm2b impairs recruitment of Ring1b to the CGIs of target genes. (a) Genome-wide occupancy of Ring1b and Ezh2 at all identified Kdm2b peaks in control (black) and Kdm2b knockdown (red) mESCs. The average signal within 5 kb genomic regions flanking the centre of Kdm2b peaks is shown. (b) Top, ChIP-seq results of Kdm2b and Ring1b at three representative genes Gata6, Sox7 and Eomes in control and Kdm2b knockdown mESCs. Schematic illustration of the genomic structure of the locus, the associated CGIs and the location of amplicons analysed by ChIP–qPCR are also shown. Bottom, the relative enrichment of Kdm2b and Ring1b occupancy at the indicated amplicons. The enrichment of occupancy was measured by ChIP–qPCR. Data are mean ± s.d., n = 3; P values (two-tailed t-test) are listed. The source data for statistics are provided in Supplementary Table S5. (c) Western blot analysis of the Ring1b, Ezh2 and Kdm2b protein levels in control and Kdm2b knockdown cells. Tubulin serves as a loading control. Full scans of blots are shown in Supplementary Fig. S8e. (d) The relative enrichment of H2A ubiquitylation levels at three representative genes (Gata6, Sox7 and Eomes) in control and Kdm2b knockdown mESCs. The enrichment was measured by ChIP–qPCR. Data are mean ± s.d., n = 3; P values (two-tailed t-test) are listed. The source data for statistics are provided in Supplementary Table S5. (e) Schematic representation of the Oct4–Sox2–Kdm2b–PRC1–CGI regulatory axis as well as the Kdm2b-dependent and the PRC2-dependent PRC1 recruitment mechanisms. The two recruitment mechanisms are likely to work in concert to silence early lineage differentiation genes in mESCs.