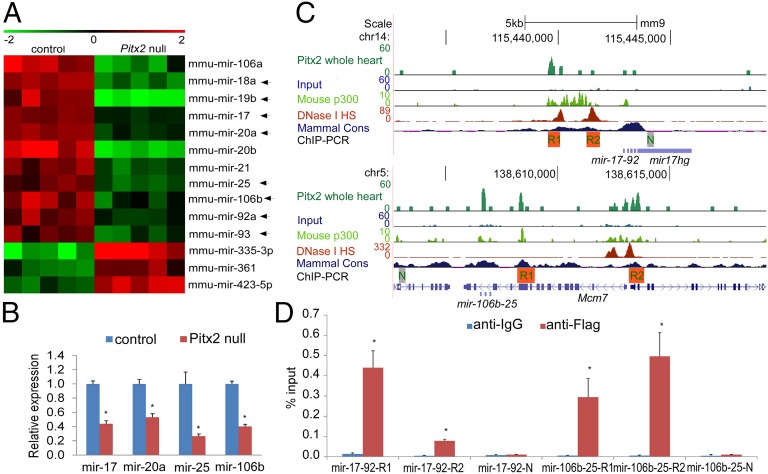
Fig. 1.
Pitx2 positively regulates miR-17-92 and miR-106b-25. (A) Heat map of miR array with wild-type control and Pitx2-null mutant hearts at E13.5. Arrows designate individual miRs from mir-17-92 and mir-106b-25. (B) Real-time PCR validation of miR array data at E13.5. *P < 0.05. (C) Three-month-old mouse whole-heart ChIP-Seq enrichment profiles (GSE50401) (18) for Pitx2-bound loci at upstream of miR-17-92 and miR-106b-25. The peaks from different datasets including Pitx2 ChIP-Seq, input track, P2 mouse heart p300 ChIP-Seq (GSE32587) (19), and 8-wk-old heart DNase I Hypersensitive Site (HS) Seq (ENCODE), as well as mammal conservation (cons) track, are aligned for comparison. Locations of real-time PCR validation of ChIP enrichment are indicated under datasets. For Pitx2 ChIP-Seq, input, and P300 ChIP-Seq, the scale indicates read count that was normalized to 10 million reads. For the DNase I HS track, we used the ENCODE default scale. N, negative control; R1, region1; R2, region 2. (D) In vivo real-time ChIP PCR indicated that miR-17-92 and miR-106b-25 were bound by Pitx2 in E13.5 mouse hearts.