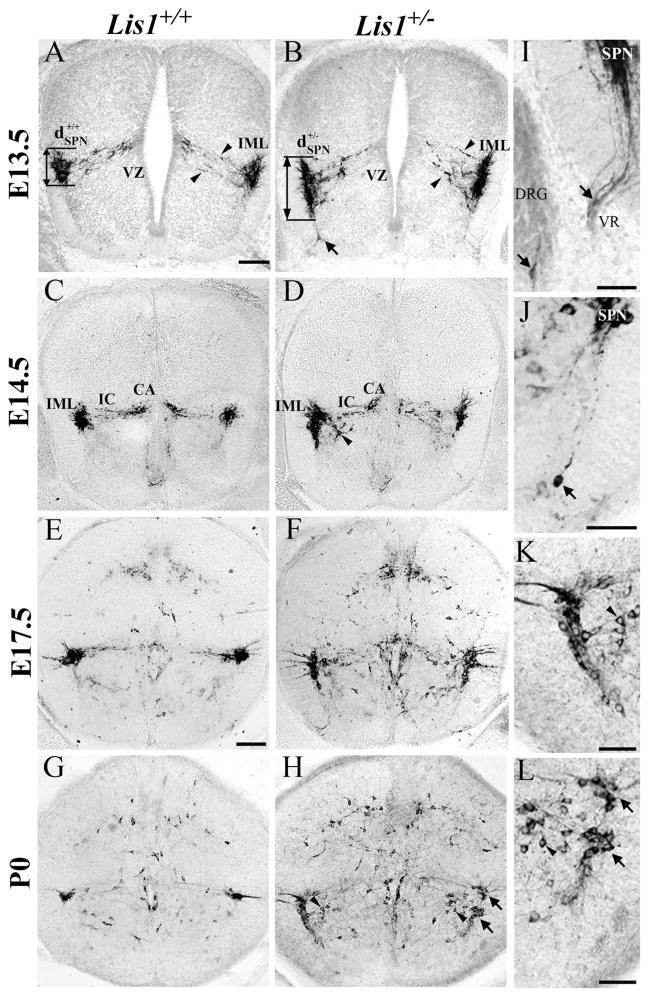
Figure 2.
Neuronal nitric oxide synthase (nNOS)-labeled Lis1+/− sympathetic preganglionic neurons (SPNs) have migratory defects. A,B: E13.5 Lis1+/+ SPNs (A, ) aggregate near the intermediolateral horn (IML), whereas Lis1+/− SPNs are distributed dorsoventrally (B, ). Lis1+/− SPNs have widespread processes (B, between arrowheads) that do not yet contact the ventricular zone (VZ) as seen in Lis1+/+ mice. Isolated nNOS-positive cell (B, arrow) is found deep in the Lis1+/− ventral horn. C–H: Most Lis1+/+ SPNs (E14.5, C; E17.5, E; P0, G) are found in the IML, and intercalated (IC) and central autonomic (CA) cells are correctly positioned. Some Lis1+/− SPNs at E14.5 (D), E17.5 (F), and P0 (H) are located in the IML whereas others are mispositioned along the tangential migratory pathway (D,F,H) or found ventromedially (H, arrowheads). I: Mispositioned E13.5 SPNs are detected in Lis1+/− dorsal root ganglion (DRG, arrow) and the ventral root (VR, arrow). J: Isolated P0 Lis1+/− SPN (arrow) is located in the deep ventral horn, widely separated from other SPNs. K,L: Enlargements of H show nNOS-labeled somata of Lis1+/− SPNs in normal (upper arrow) and ectopic (lower arrow) positions (same arrows as H). Ventromedial SPNs (arrowheads) are also mispositioned. Scale bar = 100 μm in A (applies to A–D) and E (applies to E–H); 50 μm in I–L.