Abstract
We have synthesized a 7-diethylaminocoumarin (DEAC) derivative that allows wavelength selective, two-photon uncaging at 900 nm versus 720 nm. This new caging chromophore, called DEAC450, has an extended π-electron moeity at the 3-position that shifts the absorption spectrum maximum of DEAC from 375 nm to 450 nm. Two-photon excitation at 900 nm was more than 60-fold greater than at 720 nm. Two-photon uncaging of DEAC450-Glu at 900 nm at spine heads on pyramidal neurons in acutely isolated brain slices generated postsynaptic responses that were similar to spontaneous postsynaptic excitatory miniature currents, whereas significantly higher energies at 720 nm evoked no currents. Since many nitroaromatic caged compounds are two-photon active at 720 nm, optically selective uncaging of DEAC450-caged biomolecules at 900 nm may allow facile two-color optical interrogation of bimodal signaling pathways in living tissue with high resolution for the first time.
The use and recording of color in a scientific context is now a fundamental part of what we do in biomedical research1. Thus, it is difficult to imagine using confocal fluorescent imaging if it was still monochrome2. Fortunately many technological advances have been combined to allow us to use fluorescence imaging to monitor many aspects of neuronal activity in real time in spectrally separate channels3. In contrast, our ability to manipulate cell function with comparable chromatic diversity lags behind imaging and seriously limits our ability to study multiple signaling pathways simultaneously4.
Neuroscience is a field in which optical actuation of cell function has been widely used. For example, four methods have been developed for photocontrol of neuronal membrane potential: (1) neurotransmitter uncaging to activate endogenous ligand-gated ion channels5,6; (2) chemical modification of mutated ion channels with optical switches7; (3) photochemical stimulation of genetically targeted alien ion channels8; and (4) excitation of genetically delivered photoregulated ion pumps and channels9–12. Each of these methods has advantages and disadvantages. The first is powerful because it directly activates native receptors, so is useful for understanding the details of cellular physiology in vitro13. A striking feature of methods 2 and 4 is the wavelength selectivity that is inherent to or engineered into chromophores such that two colors of light can be used orthogonally for different purposes.
Starting in 1978, hundreds of biological studies have been reported using nitrobenzylcaged compounds using photolysis at short wavelengths of light, i.e. in the 350–400 nm range for one-photon14–19 and 720–740 nm for two-photon18,20,21 photolysis. Caging chromophores that absorb at longer wavelengths than these compounds have only been recently developed, so these have been applied to relatively few biological questions. In particular, several substituted organic, laser-dye based 7-aminocoumarins22 and organicinorganic hybrid chromophores based on the ruthenium- bipyridyl23,24 (RuBi) scaffold are effectively photolyzed at wavelengths longer than 400 nm for one-photon or 740 nm for two-photon photolysis. Caged neurotransmitters using such chromophores are important additions to the optical arsenal available to neurobiologists, but their absorption spectra lack pronounced minima at short wavelengths (Figure 1). Here we introduce a new caged glutamate compound, called DEAC450-Glu (Figure 1a) that is relatively photoinactive at short wavelengths (e.g. 720 nm) and undergoes maximal two-photon excitation at 900 nm. This significant bathochromic shift thus extends the color palette of two-photon photolysis to a region that it is optically complementary to many other caged compounds.
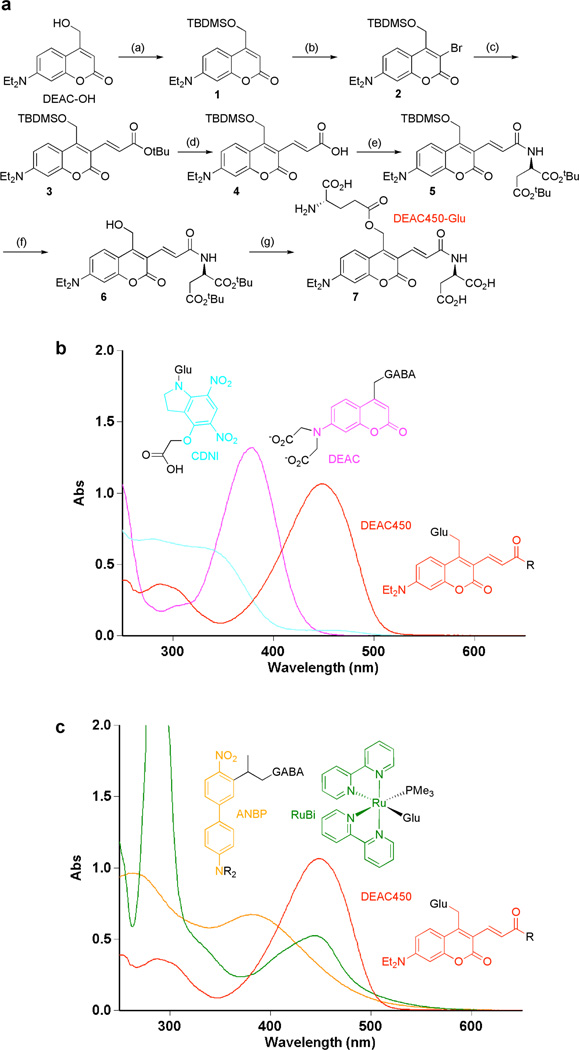
Figure 1.
Synthesis and spectral properties of DEAC450-Glu. (a) Reagents and conditions: (a) tert-Butyldimethylsilylchloride, imidazole. (b) N-bromosuccinimide, NaOAc. (c) tert-Butylacrylic acid, Pd(OAc)2, LiCl, NaHCO3, Bu4NCl. (d) TFA. (e) Di-tert-butylaspartate, EDC. (f) TBAF. (g) tert-Butyl-BOC-L-glutamate, EDC followed by TFA. (b) Absorption spectra of CDNI-Glu (cyan), N-DCAC-GABA (DEAC core in pink) and DEAC450-Glu (red). (c) Absorption spectra of RuBi-Glu (green), ANBP-GABA (orange) and DEAC450-Glu (red).
The synthesis of DEAC450-Glu (Figure 1a) started by protection of the known DEAC-alcohol25 to give 1. The coumarin 3-position was functionalized with NBS26 to give 2, followed by Heck coupling27 of tert-butylacrylic acid with 2 to give 3. Deprotection of the tert-butyl to acid 4, was followed by carbodiimide coupling of di-tert-butyl-D-Asp to give 5. Selective removal of the silyl group gave alcohol 6, which was coupled to acid side chain of L-glutamate to give fully protected DEAC450-Glu. Finally, the remaining protecting groups were removed and the product purified by HPLC to give 7 (DEAC450-Glu). DEAC450-Glu was found to be soluble (up to 7.5 mM) and quite stable at pH 7.4. Solutions showed no hydrolysis over 60 days at −20°C or at RT for 5 h, and at 37°C only 2% was hydrolyzed in 2 h. Irradiation of DEAC450-Glu and RuBi-Glu24 at 473 nm at pH 7.4 revealed that the former was photolyzed three times faster, corresponding to a quantum yield of photolysis of 0.39. DEAC450-alcohol was found, by HPLC analysis of the reaction mixture, to be cleanly released. The new chromophore has an extinction coefficient of 43,000. These data taken together show that DEAC450-Glu is photochemically one of the most efficient caged Glu probes that has been developed (Table 1). Importantly, the absorption maximum at 450 nm is significantly red-shifted compared to simple DEAC derivatives such as N-DCAC-GABA28 (Figure 1b). Furthermore, the absorption minimum of DEAC450 is at the maximum for the CDNI28,29 chromophore (Figure 1b). The relative absorption at the λmin is 9% of the λmax; no such distinct minima exist for other caged compounds (Figure 1c) that have been recently shown to be two-photon sensitive24,30,31. This relative difference in linear absorption is further enhanced in the two-photon domain; the DEAC450 chromophore is >60x more fluorescent at 900 nm than 720 nm. This significant difference in two-photon excitation lead us to test comparative uncaging of DEAC450-Glu on single spine heads in acutely isolated brain slices at these wavelengths.
Table 1.
Summary of the properties of caged glutamate probes.
| Cage | ε/M−1cm−1 (λmax, nm) |
QY | ε.QY | 2PuCS/GM (λ, nm) |
|---|---|---|---|---|
| MNI2,44 | 4,300 (330) | 0.085 | 357 | 0.06 (740) |
| RuBi24 | 5,600 (450) | 0.13 | 728 | 0.14 (800) |
| PMNB45 | 9,900 (317) | 0.1 | 990 | 0.45 (800) |
| CDNI35 | 6,400 (330) | 0.5 | 3,200 | 0.06 (720) |
| PNEB46 | 9,900 (317) | 0.1 | 990 | 3.3 (740) |
| DEAC450 | 43,000 (450) | 0.39 | 16,800 | 0.5* (900) |
Symbols and abbreviations. ε, extinction coefficient; λmax, absorption maximum; QY, quantum yield; 2PuCS, two-photon uncaging cross section;
estimated from Fig. 2.
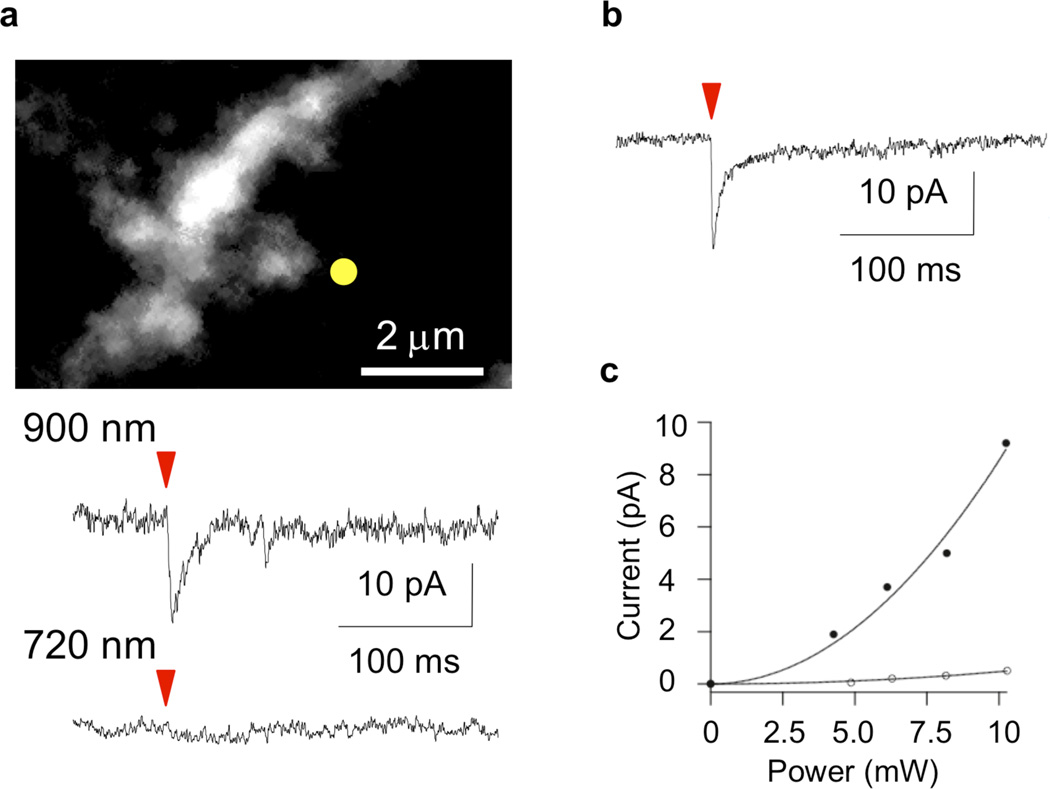
Two-photon photolysis of a solution of DEAC450-Glu (local perfusion from a nearby pipette at 0.25 mM) at 900 nm (10 mW, 0.5 ms) induced excitatory post-synaptic currents at an isolated spine head (Figure 2a) that were similar in size and duration to spontaneous miniature excitatory postsynaptic potentials32. A comparable amount (144%) of energy at 720 nm evoked no response (Figure 2a). This latter energy dose can be used to photolyze nitroindolinyl-caged neurotransmitters such as MNI-Glu32–35 (Figure 2b) and CDNI-GABA29. Note that DEAC cages and other fluorophores do not interfere with nitroaromatic photolysis at 720 nm18,28. Power response curves implied that DEAC450-Glu was uncaged by two-photon excitation at 900 nm (Figure 2c), just as MNI-Glu and NDBF-EGTA at 720 nm32,36. Similar to other caged Glu and GABA probes24,29,30, DEAC450-Glu had off-target pharmacological side effects upon GABA-A receptor currents. We found the EC50 for blocking evoked GABA-A receptor cur- rents on layer 2/3 pyramidal neurons was about 33 µM. In comparison, two commercially available caged neurotransmitters, MNI-Glu and RuBi-Glu, had EC50 values of 105 µM and 7.7 µM, respectively (Supporting Information).
Figure 2.
Optically selective two-photon uncaging of glutamate at 900 nm. Caged neurotransmitters were topically applied to pyramidal cells through a puffer pipette positioned just above the surface of an acutely isolated mouse brain slice. Excitation was with a mode-locked Ti:sapphire laser tuned to the specified wavelength. Power was measured at the exit of the microscope objective. Currents were measured at the cell soma using whole-cell voltage-clamp recordings. (a) Two-photon fluorescence image of dendritic segment on a pyramidal neuron filled with Alexa-594 (top panel). DEAC450-Glu, applied at 0.25 mM, was irradiated with 900 nm or 720 nm light for a period 0.5 ms. The photolysis beam was positioned (yellow dot) next to a spine head and the postsynaptic current induced by 10 mW at 900 nm was similar to that evoked by synaptic release. Irradiation at the same position with 12 mW of 720-nm light evoked no current. (b) MNI-Glu, applied at 10 mM, was irradiated with 720 nm light for a period of 0.5 ms and evoked a postsynaptic current that was to similar synaptic release. Importantly, nitroindolinyl-caged glutamate is 38-fold less two-photon active at 830 nm28. (c) Powerdosage curve of evoked postsynaptic current showed a two-photon excitation effect for the postsynaptic current for DEAC450-Glu at 900 nm (solid points). A much weaker response was evoked at 720 nm (open circles).
DEAC450-Glu has a uniquely powerful set of properties when compared to the many other caged glutamates. Table 1 shows a summary of the properties of a range of widely used and recently developed caged glutamate probes. It can be seen that DEAC450-Glu is highly efficient for uncaging with visible light. For example, in comparison to the two commercially available probes24,32 in Table 1, DEAC450-Glu is about 23x more active than RuBi-Glu at 450 nm, and about 329x more active than MNI-Glu at 405 nm. The latter wavelength corresponds to excitation with a purple laser that is widely deployed on confocal microscopes37. The 11-fold difference in excitation at 350 versus 450 nm may permit two-color uncaging experiments in the linear excitation domain, when DEAC450 is paired with regular nitrobenzyl caged compounds. However, we believe the real strength of our new caging chromophore for two-color uncaging may be seen when using two-photon excitation. In this modality, the relative ability to excite DEAC450 at 900 nm versus 720 nm is > 60x. Such optical selectivity allowed us to induce currents at single spine heads in acutely isolated brain slices, that were similar in size and kinetics to synaptic events32, by uncaging at 900 nm (Figure 2a). Importantly, we found that a higher energy dosage at the shorter wavelength (720 nm) evoked no significant currents (Figure 2a). This short wavelength is the one that has been widely used for uncaging nitroindolinyl-caged transmitters (e.g. MNI and CDNI compounds29,32,35, Table 1). Thus, DEAC450 and such chromophores could form a near-perfect pair of optically complementary cages for two-color, two-photon uncaging. Several other reports of two-color uncaging have appeared38–42, however all these approaches are constrained in some important way. Some require the complete photolysis of the longer wavelength before applying short wavelength uncaging38- 40. Others require the relative concentrations of the caged molecules to be set such that compounds have hugely different concentration ratios41. Finally, some chromophores require uncaging with light that is not compatible with modern microscope glass or amenable to single synapse stimulation42. In contrast, our new caging chromophore, DEAC450, enables, for the first time, two-photon uncaging of a biomolecule at long wavelengths (900 nm) with almost complete optically selectivity versus shorter wavelengths (720 nm). For synaptic physiology, this technological advance is an important break through, as it may allow the study of how excitatory and inhibitory transmitters sculpt dendritic integration with single synapse resolution43.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH (GM053395 and NS069720 to GED, NS046579 to BLS, MH099045 to MJH), and the HFSP (RG0089/2009C to GED).
Footnotes
ASSOCIATED CONTENT
Chemical and physiological experimental details. Analytical data for chemical synthesis. This material is available free of charge via the Internet at http://pubs.acs.org.”
The authors declare no competing financial interest.
REFERENCES
- 1.Miyawaki A, Sawano A, Kogure T. Nat Cell Biol. 2003;(Suppl):S1–S7. [PubMed] [Google Scholar]
- 2.Lichtman JW, Conchello JA. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki A. Neuron. 2005;48:189–199. doi: 10.1016/j.neuron.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Ellis-Davies GCR. ACS Chem Neurosci. 2011;2:185–197. doi: 10.1021/cn100111a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaway EM, Katz LC. Proc Natl Acad Sci U S A. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denk W. Proc Natl Acad Sci U S A. 1994;91:6629–6633. doi: 10.1073/pnas.91.14.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zemelman BV, Nesnas N, Lee GA, Miesenbock G. Proc Natl Acad Sci U S A. 2003;100:1352–1357. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemelman BV, Lee GA, Ng M, Miesenbock G. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 10.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 11.Han X, Boyden ES. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez VA, Sabatini BL. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 14.Ellis-Davies GCR. Nat Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer G, Heckel A. Angew Chem Int Ed Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 16.Lee HM, Larson DR, Lawrence DS. ACS Chem Biol. 2009;4:409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eder M, Zieglgansberger W, Dodt HU. Rev Neurosci. 2004;15:167–183. doi: 10.1515/revneuro.2004.15.3.167. [DOI] [PubMed] [Google Scholar]
- 18.Ellis-Davies GCR. Chem Rev. 2008;108:1603–1613. doi: 10.1021/cr078210i. [DOI] [PubMed] [Google Scholar]
- 19.Kramer RH, Chambers JJ, Trauner D. Nat Chem Biol. 2005;1:360–365. doi: 10.1038/nchembio750. [DOI] [PubMed] [Google Scholar]
- 20.Zipfel WR, Williams RM, Webb WW. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 21.Svoboda K, Yasuda R. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Givens RS, Rubina M, Wirz J. Photochem Photobiol Sci. 2012;11:472–488. doi: 10.1039/c2pp05399c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zayat L, Calero C, Albores P, Baraldo L, Etchenique R. J Am Chem Soc. 2003;125:882–883. doi: 10.1021/ja0278943. [DOI] [PubMed] [Google Scholar]
- 24.Fino E, Araya R, Peterka DS, Salierno M, Etchenique R, Yuste R. Front Neural Circuits. 2009;3:2. doi: 10.3389/neuro.04.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen V, Bendig J, Frings S, Eckardt T, Helm S, Reuter D, Kaupp UB. Angew Chem Int Ed Engl. 2001;40:1045–1048. doi: 10.1002/1521-3773(20010316)40:6<1045::aid-anie10450>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 26.Guo HM, Tanaka F. J Org Chem. 2009;74:2417–2424. doi: 10.1021/jo900013w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pottie IR, Nandaluru PR, Benoit WL, Miller DO, Dawe LN, Bodwell GJ. J Org Chem. 2011;76:9015–9030. doi: 10.1021/jo201775e. [DOI] [PubMed] [Google Scholar]
- 28.Kantevari S, Matsuzaki M, Kanemoto Y, Kasai H, Ellis-Davies GCR. Nat Methods. 2010;7:123–125. doi: 10.1038/nmeth.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuzaki M, Hayama T, Kasai H, Ellis-Davies GCR. Nat Chem Biol. 2010;6:255–257. doi: 10.1038/nchembio.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donato L, Mourot A, Davenport CM, Herbivo C, Warther D, Leonard J, Bolze F, Nicoud JF, Kramer RH, Goeldner M, Specht A. Angew Chem Int Ed Engl. 2012;51:1840–1843. doi: 10.1002/anie.201106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gug S, Charon S, Specht A, Alarcon K, Ogden D, Zietz B, Leonard J, Haacke S, Bolze F, Nicoud JF, Goeldner M. Chembiochem. 2008;9:1303–1307. doi: 10.1002/cbic.200700651. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki M, Ellis-Davies GCR, Nemoto T, Miyashita Y, Iino M, Kasai H. Nat Neurosci. 2001;4:1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higley MJ, Sabatini BL. Neuron. 2008;59:902–913. doi: 10.1016/j.neuron.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Kwon HB, Sabatini BL. Nature. 2011;474:100–104. doi: 10.1038/nature09986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis-Davies GCR, Matsuzaki M, Paukert M, Kasai H, Bergles DE. J Neurosci. 2007;27:6601–6604. doi: 10.1523/JNEUROSCI.1519-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momotake A, Lindegger N, Niggli E, Barsotti RJ, Ellis-Davies GCR. Nat Methods. 2006;3:35–40. doi: 10.1038/nmeth821. [DOI] [PubMed] [Google Scholar]
- 37.Trigo FF, Corrie JE, Ogden D. J Neurosci Methods. 2009;180:9–21. doi: 10.1016/j.jneumeth.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 38.Goguen BN, Aemissegger A, Imperiali B. J Am Chem Soc. 2011;133:11038–11041. doi: 10.1021/ja2028074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotzur N, Briand B, Beyermann M, Hagen V. J Am Chem Soc. 2009;131:16927–16931. doi: 10.1021/ja907287n. [DOI] [PubMed] [Google Scholar]
- 40.Priestman MA, Sun L, Lawrence DS. ACS Chem Biol. 2011;6:377–384. doi: 10.1021/cb100398e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.San Miguel V, Bochet CG, del Campo A. J Am Chem Soc. 2011;133:5380–5388. doi: 10.1021/ja110572j. [DOI] [PubMed] [Google Scholar]
- 42.Stanton-Humphreys MN, Taylor RD, McDougall C, Hart ML, Brown CT, Emptage NJ, Conway SJ. Chem Commun (Camb) 2012;48:657–659. doi: 10.1039/c1cc15135e. [DOI] [PubMed] [Google Scholar]
- 43.Gidon A, Segev I. Neuron. 2012;75:330–341. doi: 10.1016/j.neuron.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Papageorgiou G, Corrie JET. Tetrahedron. 2000;56:8197–8205. [Google Scholar]
- 45.Specht A, Thomann JS, Alarcon K, Wittayanan W, Ogden D, Furuta T, Kurakawa Y, Goeldner M. Chembiochem. 2006;7:1690–1695. doi: 10.1002/cbic.200600111. [DOI] [PubMed] [Google Scholar]
- 46.Specht A, Bolze F, Donato L, Herbivo C, Charon S, Warther D, Gug S, Nicoud JF, Goeldner M. Photochem Photobiol Sci. 2012;11:578–586. doi: 10.1039/c2pp05360h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.