Abstract
Bacteria of the genus Xenorhabdus are symbionts of soil entomopathogenic nematodes of the genus Steinernema. This symbiotic association constitutes an insecticidal complex active against a wide range of insect pests. Unlike other Xenorhabdus species, Xenorhabdus poinarii is avirulent when injected into insects in the absence of its nematode host. We sequenced the genome of the X. poinarii strain G6 and the closely related but virulent X. doucetiae strain FRM16. G6 had a smaller genome (500–700 kb smaller) than virulent Xenorhabdus strains and lacked genes encoding potential virulence factors (hemolysins, type 5 secretion systems, enzymes involved in the synthesis of secondary metabolites, and toxin–antitoxin systems). The genomes of all the X. poinarii strains analyzed here had a similar small size. We did not observe the accumulation of pseudogenes, insertion sequences or decrease in coding density usually seen as a sign of genomic erosion driven by genetic drift in host-adapted bacteria. Instead, genome reduction of X. poinarii seems to have been mediated by the excision of genomic blocks from the flexible genome, as reported for the genomes of attenuated free pathogenic bacteria and some facultative mutualistic bacteria growing exclusively within hosts. This evolutionary pathway probably reflects the adaptation of X. poinarii to specific host.
Keywords: entomopathogenic bacteria, Lepidoptera, Steinernema, comparative genomics, regions of genomic plasticity, genomic deletion
Introduction
Symbioses between microorganisms and animals are widespread in numerous ecological niches. Mutualistic symbiosis is based on mutual exploitation, in which each organism contributes to the interaction but receives a net positive benefit. The benefits are diverse and include a mutual influence on nutrition, defense, reproduction, and development (Chaston and Goodrich-Blair 2010). Multipartite microbial symbiosis involves long-term associations between three or more species, with at least two of the partners benefiting from the interaction (Hussa and Goodrich-Blair 2013). In the Steinernema–Xenorhabdus symbiotic interaction, soil entomopathogenic nematodes from the genus Steinernema are dependent on their intestinal bacterial symbiont, Xenorhabdus (Enterobacteriaceae), for colonization of the insects serving as their nutritional and reproductive niche. A nonfeeding soil-dwelling infective juvenile stage of the nematode penetrates the hemocel of the insect and releases the bacteria into the hemolymph. The bacterial symbiont helps to overcome insect immunity, kills the insect, and converts the cadaver into an essential source of food for nematode growth and development. Nematodes undergo several rounds of reproduction within the insect cadaver. When nematode density becomes too high and the nutrients derived from the cadaver are exhausted, the bacteria recolonize the nematodes, which then emerge from the insect cadaver into the soil, to search for a new host (Goodrich-Blair and Clarke 2007; Richards and Goodrich-Blair 2009; Nielsen-LeRoux et al. 2012). No free-living forms of Xenorhabdus have ever been isolated outside of the nematode host (Forst et al. 1997). Except the direct vectorization in the insect hemolymph, the benefit of the association to the bacterium has yet to be elucidated.
Since the 1980s, various species of entomopathogenic nematodes have been sold and used as effective biological control. agents for soil-inhabiting insects. Field and laboratory studies have demonstrated the importance of matching the appropriate nematode species with the particular pest targeted (Ehlers 2001). For example, the Steinernema carpocapsae–Xenorhabdus nematophila couple is virulent in various insect orders, and has been shown to be effective against Pseudaletia unipuncta (Lepidoptera), Acheta domesticus (Orthoptera), and Plectrodera scalator (Coleoptera). By contrast, the St. glaseri–Xenorhabdus poinarii couple is virulent principally in a few coleopteran species, such as Popillia japonica and Cyclocephala hirta, suggesting a potentially narrow host specificity (Wang et al. 1994; Converse and Grewal 1998; Rosa et al. 2002; Fallon et al. 2006).
Co-operation between the bacterial and the helminthic partner also differs between entomopathogenic couples. Under laboratory conditions, the bacterium or the nematode can be entomopathogenic alone. The injection of a dose of 100 cells of the bacteria X. nematophila and X. bovienii into larvae is lethal in diverse insects (Poinar and Thomas 1966; Forst et al. 1997; Ansari et al. 2003; Sugar et al. 2012). Aposymbiotic St. carpocapsae and St. feltiae nematodes lacking the Xenorhabdus symbiont can kill Galleria mellonella or Tipula oleracea although nematode reproduction is less efficient in the insect cadaver in the absence of the symbiont (Ehlers et al. 1997; Han and Ehlers 2000). These associations can, therefore, be considered as facultative for both partners under laboratory conditions. For example, a substantial body of molecular data has been accumulated on the factors enabling bacteria of species X. nematophila to adapt to the insect host in the absence of the nematode (Herbert et al. 2007; Nielsen-LeRoux et al. 2012). By contrast, co-operation is of much greater importance for the killing of insects in some bacterium-nematode complexes. In the St. glaseri–X. poinarii couple, the bacterial symbiont is avirulent or only weakly virulent when artificially injected into several insects (Akhurst 1986; Rosa et al. 2002; Ansari et al. 2003). No mortality is observed after the experimental infestation of G. mellonella with axenic St. glaseri nematodes (Akhurst 1986). However, a large proportion of St. glaseri nematodes are naturally aposymbiotic (Akhurst 1986). These contradictory features make it difficult to evaluate the facultative status of the St. glaseri–X. poinarii association.
Host-adapted bacteria have been described in both mutualistic and pathogenic symbioses. Obligatory mutualistic symbionts of insects (e.g., Buchnera, Wigglesworthia, etc.) live in specialized host organs (bacteriomes) and share a long-standing coevolutionary history with their host. They are vertically transmitted and display extreme genomic reduction. Facultative mutualistic symbionts are not strictly necessary for their host. They do not live exclusively in a specialized organ and undergo horizontal transfers between host strains or species (Dale and Moran 2006). These symbionts may be found in an active free-living stage (e.g., the squid symbiont Vibrio fischeri) or may grow exclusively within the hosts (e.g., the insect symbiont Wolbachia). Wolbachia, like some host-adapted pathogenic bacterial species, such as Burkholderia mallei and Mycobacterium leprae, displays a massive expansion of insertion sequences (IS), leading to pseudogene formation, chromosomal rearrangements mediated by recombination between IS and moderate genome downsizing. These features are considered to constitute the initial stages of a drastic reduction of genome size (Moran and Plague 2004; Gomez-Valero et al. 2007; Song et al. 2010). Finally, some host-adapted pathogenic bacteria, such as My. tuberculosis and the asymptomatic bacteriuria (ABU) strains of Escherichia coli, display moderate genome downsizing without massive IS expansion (Zdziarski et al. 2008; Veyrier et al. 2011). In such cases, the decrease in genome size probably results from the excision of mobile genetic elements.
We focused here on the pathogenic and genomic properties of the very poorly documented species X. poinarii. We compared the virulence of five X. poinarii strains by injecting them into the lepidopteran Spodoptera littoralis. We confirmed that all five strains tested had attenuated virulence. We sequenced the genomes of the X. poinarii G6 strain (Xp_G6) and the X. doucetiae FRM16 (Xd) strain, a closely related virulent strain. We showed that the genome of Xp_G6 was much smaller (500–700 kb) than those of Xd, X. nematophila ATCC19061 (Xn) and X. bovienii SS-2004 (Xb), which have recently been sequenced and analyzed (Ogier et al. 2010; Chaston et al. 2011). This small genomic size is a key feature of the X. poinarii species resulting from reductive evolution, probably mediated by the deletion of regions of genomic plasticity (RGP). Thus, our study made it possible to compare the evolutionary history of the X. poinarii genome with that of other Xenorhabdus species.
Materials and Methods
Bacterial Strains, Media, Phenotypic Analyses, and Genomic DNA Extraction
All the bacterial strains used in this study are listed in table 1. Xenorhabdus strains were routinely grown in Luria–Bertani (LB) broth, 1.5% nutrient agar medium (GNO) or NBTA medium (GNO supplemented with 25 mg bromothymol blue and 40 mg triphenyl-2,3,4 tetrazolium chloride per liter) at 28 °C. Bacteria were stored at −80 °C in 16% glycerol (v/v). Genomic DNA was extracted as previously described (Gaudriault et al. 2006) and stored at 4 °C.
Table 1.
List of Xenorhabdus Strains Used in This Study
| Strain | Species | Nematode Host from Which Strain Was Isolated | Geographical Origin | Reference |
|---|---|---|---|---|
| G6 (ATCC 49121) | X. poinarii | Steinernema glaseri | USA (NC) | Akhurst 1986 |
| AZ26 | X. poinarii | St. glaseri | Portugal | Rosa et al. 2002 |
| NC33 | X. poinarii | St. glaseri | USA (NC) | Fischer-Le Saux, Arteaga-Hernandez, et al. 1999 |
| SK72 | X. poinarii | St. glaseri | USA (FL) | Fischer-Le Saux, Arteaga-Hernandez, et al. 1999 |
| CU01 | X. poinarii | St. cubanum | Cuba | Fischer-Le Saux, Arteaga-Hernandez, et al. 1999 |
| ATCC19061 | X. nematophila | St. carpocapsae | USA | Chaston et al. 2011 |
| SS2004 | X. bovienii | St. jolietti | USA (MO) | Chaston et al. 2011 |
| FRM16 | X. doucetiae | St. diaprepesi | Martinique | Fischer-Le Saux, Arteaga-Hernandez, et al. 1999 |
Phylogenetic Analysis
Sequence alignment was generated and phylogenetic methods were performed as previously described (Tailliez et al. 2010, 2012). Briefly, for each bacterial strain, individual gene fragments (recA, 646 nucleotides; gyrB, 864 nucleotides; dnaN, 828 nucleotides; gltX, 913 nucleotides; and infB, 965 nucleotides) were aligned using MUSCLE (Edgar 2004) and then concatenated with the seaview platform (http://doua.prabi.fr/software/seaview, last accessed June 17, 2014). Ambiguously aligned blocks were removed by the Gblocks method (Castresana 2000) or the “Guidance” program (Penn et al. 2010). Maximum-likelihood analysis (phyML 3.0) was carried out with the general time-reversible model of substitution with gamma-distributed rate heterogeneity and a proportion of invariant sites determined for all five protein-coding sequences by jModelTest, to give the best fit to the data according the Akaike information criterion (Posada and Crandall 1998). MUSCLE, Gblocks, PhyML, and bootstrap values were obtained from the phylogeny.fr platform (Dereeper et al. 2008). Five X. poinarii strains, 23 type strains representative of Xenorhabdus species and the Xb strain, the genome of which has been sequenced, were included in this study. Three strains of Photorhabdus and one strain of Proteus mirabilis were used as closely related outgroups. The accession numbers of the individual genes used for building phylogenetic trees are listed in supplementary table S1, Supplementary Material online. The Enterobacteriaceae phylogenetic tree was constructed as described above, from the concatenated sequences of 12 conserved individual genes from the core genome (infB, nusA, polA, pyrD, rpoB, valS, cysS, metK, purA, tpiA, smpB, secY) of 47 Enterobacteriaceae strains. These genes belong to the defined set of 205 single-copy genes resistant to horizontal genetic transfer (HGT) and providing a reliable and consistent reconstruction of the phylogeny of γ-Proteobacteria (Lerat et al. 2003). These 12 genes were chosen for study on the basis of their homogeneous distribution along the length of the chromosome, and their similar model of DNA sequence evolution, as assessed with jModeltest (Posada and Crandall 1998). The nucleotide sequences used to construct the phylogenetic trees for Enterobacteriacae and xaxA were extracted from publicly available genomes.
Insect Pathogenicity Assays
Bacteria were directly injected into two model insects: Sp. littoralis (Lepidoptera: Noctuidae) corn variant from Spain and G. mellonella (Lepidoptera: Pyralidae), as previously described (Sicard et al. 2004). For Sp. littoralis, all injections were performed on 1-day-old sixth-instar larvae that had been reared on an artificial diet (Poitout 1970) at 23 ± 1 °C, with a photoperiod of L16:D8 and a relative humidity of 40 ± 5%. For G. mellonella, all injections were performed on last-instar larvae reared at 28 °C in the dark with honey and pollen. Xenorhabdus strains were grown in LB broth (Difco) at 28 °C, with shaking, to exponential growth phase, corresponding to an optical density of 0.8 at 600 nm (Jenway Colorimeter). We injected 20 µl of bacterial suspension, containing 500–1,000 cells, into 20 larvae, with a Hamilton syringe. The surface of the insect larva was sterilized with 70% (v/v) ethanol before the injection of the bacteria into the hemocoel. The number of bacteria injected into the larvae was checked by plating serial dilutions on LB agar plates. Insect mortality was assessed at regular point times after injection, for the evaluation of LT50. At least four independent experiments were performed for each strain. For Sp. littoralis assays, statistical analysis was carried out with the Statistical Package for Social Science version 11.0.1 (SPSS, Chicago, IL), comparing individual survival times within each group.
Sequencing and Assembly of the Whole Genomes
The complete genome sequences of Xp_G6 and Xd were obtained by a mixture of Sanger capillary and new sequencing technologies. We first added about 23-fold coverage of 454 GSflx (Roche; www.roche.com, last accessed June 17, 2014) reads to Sanger reads, derived from a library with an insert fragment size of 10 kb. This library was constructed by the mechanical shearing of genomic DNA and insertion of the fragments generated into pCNS (pSU18-derived). Plasmid DNA was then purified and end-sequenced (5,745 reads for Xp_G6 and 5,877 reads for Xd) by dye-terminator techniques, with ABI3730 sequencers (Applied Biosystems, Foster City, CA), resulting in approximately 1-fold coverage for both genomes. After assembly with Arachne (www.broadinstitute.org, last accessed June 17, 2014), to decrease the number of scaffolds, 5.6-fold coverage mate-paired 454 GSflx reads (with a library insert size of about 3 kb) were then added. The whole reads were assembled with Newbler (Roche) and validated via the Consed interface (www.phrap.org, last accessed June 17, 2014). For the finishing phase, we used primer walking on clones, polymerase chain reaction (PCR) and in vitro transposition technology (Template Generation System II Kit; Finnzyme, Espoo, Finland), generating 359, 162 and 533 additional reads, respectively, for Xp_G6 and 701, 823 and 3,701 additional reads, respectively, for Xd. Illumina reads (36 bp) corresponding to a coverage of about 50-fold were mapped with SOAP (http://soap.genomics.org.cn, last accessed June 17, 2014) during the polishing phase, as previously described (Aury et al. 2008).
PCR Amplification and the Sequencing of Nucleotidic Fragments
PCR amplifications targeting selected genomic regions were carried out, for analysis of the distributions of these regions in a panel of Xenorhabdus strains. Consensual pairs of primers (supplementary table S2, Supplementary Material online) were manually designed from clustalW alignments (http://multalin.toulouse.inra.fr/multalin/, last accessed June 17, 2014) of selected regions of Xenorhabdus reference genomes. Fragments with a predicted size of less than or greater than 3 kb were amplified with Taq polymerase (Invitrogen) or with the High Proof DNA Polymerase (BioRad), respectively, according to the manufacturer’s protocol. PCR amplifications were performed with a BioRad thermocycler (BioRad), and PCR products were analyzed by electrophoresis in an agarose gel. Amplicons were sequenced by MWG-Eurofins France.
Pulsed-Field Gel Electrophoresis Analysis
Intact genomic DNA was extracted in agarose plugs as follows. Bacterial cells grown on nutrient agar plates were suspended in phosphate-buffered saline (GIBCO; Invitrogen) to a turbidity of 1.25 at 650 nm, included in 1.2% low-melting point agarose (SeaPlaque®GTG) solution (v/v) and lysed, as previously described (Jumas-Bilak et al. 1998). I-CeuI (New England Biolabs) hydrolysis was performed as previously described (Teyssier et al. 2005). Pulsed-field gel electrophoresis (PFGE) was performed in a 0.8% agarose gel, in 0.5× Tris–borate–ethylenediaminetetraacetic acid buffer, at 4.5 V/cm and 10 °C, in a CHEF-DRII apparatus (BioRad). The separation of I-CeuI fragments was optimized by using different electrophoresis conditions for fragments of different sizes: 1) a pulse ramp from 5 to 35 s for 24 h for fragments of less than 500 kb in size and 2) a pulse ramp from 150 to 400 s for 45 h for I-CeuI fragments between 500 and 4,000 kb in size. The molecular markers used were the chromosomes of Saccharomyces cerevisiae and Hansenula wingei (BioRad).
Genomic Analyses
Genome Annotation
Functional annotation was carried out with the tools of the MicroScope platform (Vallenet et al. 2013) and the annotated genome was implemented in the public XenorhabduScope database (https://www.genoscope.cns.fr/agc/microscope/home/index.php, last accessed June 17, 2014). We used specific tools for the annotation of specific gene families. The nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) genes were predicted by the “2metDB” method (Bachmann and Ravel 2009) implemented at the MicroScope platform. Toxin–antitoxin (TA) systems were predicted with RASTA-Bacteria software (Sevin and Barloy-Hubler 2007) and we focused on predicted proteins with a domain characteristic of one of the nine most frequent TA system families: CcdA/CcdB, HicA/HicB, HigB/HigA, HipA/HipB, MazE/MazF, ParD/ParE, Phd/Doc, RelB/RelE, and VapB/VapC (Pandey and Gerdes 2005). We used the ISsaga platform (http://www-is.biotoul.fr/, last accessed June 17, 2014) to count IS (Varani et al. 2011).
Synteny Analysis
Whole-genome alignments were performed with the “Synteny Line Plot” tool available from the MaGe Platform (http://www.genoscope.cns.fr/agc/mage, last accessed June 17, 2014), which carries out a global comparison of two bacterial genomes on the basis of synteny results. The percentage of coding sequences (CDS) displaying synteny between the four genomes was calculated with the synteny statistic tool available from the MaGe Platform. The minimum size of the synteny groups was five genes.
Core and Flexible Genome Analysis
We used the SiLiX program of the MicroScope platform to cluster proteins into families of homologous sequences (MICFAM) (Miele et al. 2011). This program computes pan, core, and flexible genomes.
Analysis of Mobile Genetic Elements
RGP were sought in the four Xenorhabdus genomes (except for the plasmid of Xn). First, Prophinder was first used to detect prophages in the four Xenorhabdus genomes (Lima-Mendez et al. 2008) (http://aclame.ulb.ac.be/Tools/Prophinder/, last accessed June 17, 2014). We then used the RGPFinder web tool implemented via the MaGe annotation platform (http://www.genoscope.cns.fr/agc/mage, last accessed June 17, 2014) to identify GI (genomic islands) and RGPsensu stricto (see Ogier et al. [2010] for detailed procedure). Briefly, RGPFinder searches for genomic regions (minimal size of 5 kb) displaying breaks in synteny between a query genome and a set of closely related genomes. If the regions displayed characteristics typical of foreign DNA acquired by HGT, such as compositional bias (GC% deviation, codon adaptation index) or tRNA, IS, integrase genes and genetic elements involved in DNA mobility, they were classified as GI. Regions without such features were classified as RGPsensu stricto. For the identification of integrative and conjugative elements (ICEs) in the Xenorhabdus genomes, we searched for genes encoding conjugation machinery, which consists of a relaxase, a T4SS and a type 4 coupling protein (Guglielmini et al. 2011). The ICE core was completed by searches for genes involved in 1) ICE replication, 2) DNA integration/excision, and 3) pilus biosynthesis (Seth-Smith et al. 2012).
Comparison of Gene Content
We used the MicroScope Gene Phyloprofile tool to identify sets of genes specific to Xenorhabdus genomes, with the following homology constraints: bidirectional best hit, minimal alignment coverage of 0.8, and amino acid sequence identity of 30%.
Gene Remnant Identification
We analyzed gene remnants in the Xp_G6 genome by first extracting protein sequences from the Xd genome present in the other two virulent strains, Xn and Xb, but absent from Xp_G6 using results of the Gene Phyloprofile tool from the MicroScope platform and a custom-designed Perl Script. We then compared each of these proteins with the six-frame translations of the complete genome of Xp_G6, using the TBLASTN software, a sensitive method of searching for traces of partial coding regions not annotated in the Xp_G6 genome. A gene was considered to be remnant in the Xp_G6 genome if the corresponding TBLASTN results met the following criteria: HSP (high-scoring segment pair), including one to three different hits displaying at least 40% identity and with an e value <0.01.
Results
Phylogenetic Features of the Species X. poinarii
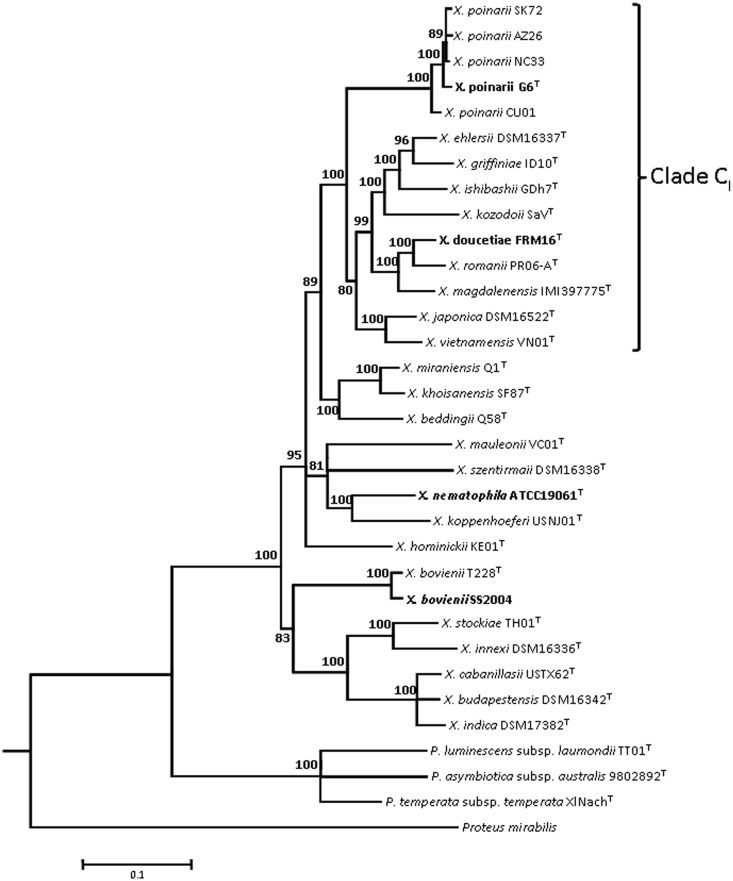
Strains AZ26, CU01, G6, NC33, and SK72 were previously classified within the species X. poinarii (Fischer-Le Saux, Viallard, et al. 1999; Tailliez et al. 2006) (table 1). We determined the phylogenetic position of the five strains within the genus Xenorhabdus with five concatenated protein-coding sequences: recA, gyrB, dnaN, gltX, and infB (fig. 1). The five strains clustered together on a clearly separate subbranch of clade CI (Tailliez et al. 2010, 2012). Therefore, the species X. poinarii emerged from the clade CI ancestor. The CU01 strain, which was located in a slightly different position, could be seen as lying at the edge of the species X. poinarii, as previously suggested by other authors (Fischer-Le Saux, Arteaga-Hernandez, et al. 1999; Tailliez et al. 2006).
Fig. 1.—
Maximum-likelihood phylogenetic tree showing the positions of Xenorhabdus poinarii strains within the genus Xenorhabdus. The analysis is based on five concatenated protein-coding sequences (recA, gyrB, dnaN, gltX, and infB). It was carried out with the GTR model of substitution, with a gamma-distributed rate heterogeneity and a proportion of invariant sites. Photorhabdus and Proteus sequences were used as outgroups. Bootstrap values (Felsenstein 1988) of more than 80% (from 100 replicates) are indicated at the nodes. Clade CI, which includes all the X. poinarii strains, is as previously described (Tailliez et al. 2010). The names of strains for which genomes have previously been sequenced or were sequenced in this study are indicated in bold italic and bold normal typescript, respectively. Bar: 10% divergence.
Pathology of the Species X. poinarii
The species X. poinarii has been described as weakly virulent following its direct injection into insect hemolymph (Akhurst 1986; Converse and Grewal 1998; Rosa et al. 2002; Ansari et al. 2003). We investigated the virulence of X. poinarii strains AZ26 (Xp_AZ26), CU01 (Xp_CU01), Xp_G6, NC33 (Xp_NC33) and SK72 (Xp_SK72) in insect larvae, by injecting 1,000 bacterial cells/larva directly into the hemocoel of Sp. littoralis and G. mellonella, these two insects being highly resistant and susceptible, respectively, to pathogenic bacteria. As a control, we used the strains Xn and Xb, two virulent Xenorhabdus reference strains, the genomes of which have been sequenced (Chaston et al. 2011). The two reference strains rapidly killed Sp. littoralis larvae, with an LT50 of 24–26 h, whereas the X. poinarii strains were strictly nonpathogenic (P < 0.05; table 2). By contrast, in G. mellonella, three X. poinarii strains (Xp_G6, Xp_SK72, and Xp_CU01) were found to be as virulent as the reference strains Xn and Xb (LT50 < 21 h). Xp_NC33 was slightly attenuated (LT50 = 35 h) and Xp_AZ26 was strictly nonpathogenic (table 2). We checked that weak virulence was not a feature common to the strains of phylogenetic clade CI, by also investigating the virulence of a type strain of this clade, Xd. This strain was as virulent as the reference strains, Xn and Xb in both insects (P < 0.05; table 2). In conclusion, the avirulence of the X. poinarii species in the highly resistant insect, Sp. littoralis, is a feature specific to this species.
Table 2.
Pathogenicity of Eight Bacterial Strains Tested by Intrahemocoelic Injection into Last Instar-Larvae of Spodoptera littoralis (incubated at 23 °C) and Galleria mellonella (incubated at 28 °C)
| Species | Strain | LT50 (h)a |
|
|---|---|---|---|
| Sp. littoralisb | G. mellonella | ||
| Xenorhabdus poinarii | G6 | No mortality | <21 |
| X. poinarii | AZ26 | No mortality | No mortality |
| X. poinarii | NC33 | No mortality | 35 |
| X. poinarii | SK72 | No mortality | <21 |
| X. poinarii | CU01 | No mortality | <21 |
| X. nematophila | ATCC19061 | 24 | <21 |
| X. bovienii | SS-2004 | 25–26 | <21 |
| X. doucetiae | FRM16 | 26–27 | <21 |
aMortality was recorded over 48 h; intrahemocoel injection of 1,000 bacterial cells/larva.
bStatistical analysis was carried out for each strain on at least four independent experiments (P < 0.05).
Sequencing of the Xp_G6 and Xd Genome Sequences
We investigated the genomic content of X. poinarii by sequencing the genome of Xp_G6, the type strain of the species isolated from the nematode St. glaseri G6 in North Carolina (Akhurst 1982, 1983; Akhurst and Boemare 1988). We also sequenced the genome of Xd, for a comparison of the genome of Xp_G6 with a closely related pathogenic Xenorhabdus strain.
General Genome Features
The genomes of Xp_G6 and Xd consist of circular chromosomes of 3,659,523 and 4,195,202 bp, encoding 3,715 and 3,974 proteins, respectively. In addition, Xd also harbors an 8,449-bp plasmid containing 12 protein-coding sequences displaying little similarity to the other CDS described, except for a putative sugar fermentation stimulation protein B (Ner-like protein) and a putative ParDE TA system (supplementary table S3, Supplementary Material online). The Xp_G6 chromosome is clearly smaller (from 536 to 773 kb smaller) than the chromosome of the three virulent strains Xn, Xb, and Xd (table 3). The Xp_G6 chromosome harbors fewer pseudogenes than the chromosomes of Xn, Xb and Xd, and this difference was particularly marked for the comparison with the 4.4 Mb chromosome of Xn, which is particularly rich in pseudogenes. It also has fewer repeated regions that usually serve as a substrate for chromosomal deletions and rearrangements (Treangen et al. 2009) than the chromosome of Xn, Xb, and Xd. The four strains have similar coding sequence densities (from 80% to 86%). Xp_G6 and Xd contain 156 and 192 putative IS, respectively, a much smaller number than for Xn and Xb (436 and 369, respectively).
Table 3.
Comparison of the Genomic Features in Xenorhabdus nematophila ATCC19061, X. bovienii SS-2004, X. doucetiae FRM16, and X. poinarii G6
| Feature |
X. nematophila ATCC19061 |
X. bovienii SS-2004 |
X. doucetiae FRM16 |
X. poinarii G6 | ||
|---|---|---|---|---|---|---|
| Chromosome | Plasmid | Chromosome | Chromosome | Plasmid | Chromosome | |
| Size (bp) | 4,432,590 | 155,327 | 4,225,498 | 4,195,202 | 8,449 | 3,659,522 |
| G+C content (%) | 44.19 | 45.97 | 44.97 | 45.71 | 45.09 | 44.55 |
| CDS | 4,299 | 175 | 4,260 | 3,974 | 12 | 3,715 |
| Coding density (%) | 80.52 | 79.62 | 85.64 | 85.93 | 50.21% | 84.67 |
| Mean CDS (bp) | 860 | 711 | 850 | 933 | 464 | 856 |
| Mean intergenic length (bp) | 163 | 150 | 158 | 155 | 495 | 164 |
| Repeated regions (%) | 16.58% | 17.99% | 16.27% | 16.18% | 0 | 9.21% |
| Pseudogenes | 99 | 4 | 58 | 45 | 0 | 38 |
| IS | 436 | ND | 369 | 192 | ND | 156 |
| Phage genes | 275 (6) | ND | 437 (8) | 157 (7) | 0 | 377 (7) |
| rRNA operons | 7 | 0 | 7 | 7 | 0 | 7 |
| tRNAs | 79 | 0 | 83 | 76 | 0 | 77 |
| Accession number | FN667742 | FN667743 | FN667741 | FO704550 | FO704549 | FO704551 |
Note.—ND, not determined.
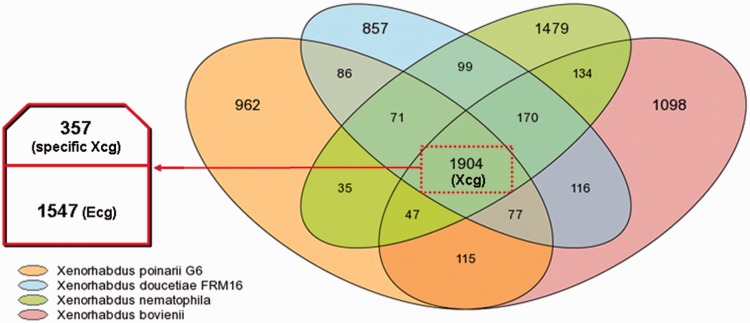
Xenorhabdus Pan Genome, Core Genome, and Flexible Genome
We analyzed the pan, core, and flexible genomes of the four Xenorhabdus genomes (fig. 2). The Xenorhabdus pan genome, corresponding to the total number of gene families present in Xenorhabdus, consists of 7,250 gene families. The Xenorhabdus core genome (Xcg), corresponding to the set of gene families common to the four Xenorhabdus strains, consisted of 1,904 gene families, or 40–50% of all the gene families present in each Xenorhabdus strains. We then introduced Es. coli strain K12, a commensal Enterobacteriaceae strain, into the analysis, which made it possible to identify an Enterobacteriaceae core genome (Ecg) of 1,547 gene families. The subtraction of the Ecg from the Xcg left us with 357 gene families that we considered to constitute the specific Xcg (see list in supplementary table S4, Supplementary Material online). The specific Xcg probably includes genes encoding factors essential for the Xenorhabdus lifestyle, particularly for symbiosis with the Steinernema and pathogenicity in insects. It encompasses many gene families previously described as encoding putative effectors of host interactions in the highly studied species X. nematophila: 1) factors potentially involved in hemocyte toxicity, such as the XhlA hemolysin, the RtxA toxin, the pore-forming fimbrial subunit MrxA, and enzymes involved in the synthesis of the lipopolysaccharide endotoxin; 2) enzymes required for the biosynthesis of rhabduscin, which inhibits phenoloxidase activity, an innate immune defense strategy of insects; 3) PrtS, PrtA, XlpA, EstA, PulA, extracellular enzymes probably involved in cadaver degradation; and 4) xenorhabdicin, a phage tail-like bacteriocin involved in intraspecies and interspecies competition within the nematode partner (Herbert et al. 2007; Chaston et al. 2011; Crawford et al. 2012). Interestingly, in addition to many genes encoding proteins of unknown functions, the specific Xcg also contains genes potentially involved in iron metabolism and transport, sodium transport, histidine and thiamine metabolism, and resistance to tellurium.
Fig. 2.—
Venn diagram showing numbers of orthologous genes in the genomes of Xenorhabdus nematophila ATCC19061, X. bovienii SS-2004, X. doucetiae FRM16 and X. poinarii G6. The Xcg (1,904 gene families) is framed by red dashes, and includes the Ecg (1,547 gene families common to Escherichia coli K12) and the specific Xcg (357 gene families).
The flexible genome (corresponding to the subtraction of the Xcg from the pan genome) consists of gene families absent from at least one of the genomes compared. The flexible genome of each strain accounts for 42%, 44%, 53% and 49% of the total numbers of gene families in the Xp_G6, Xd, Xn and Xb genomes, respectively. The flexible genome is rich in strain-specific gene families (23–36%) mostly annotated as conserved genes of unknown function, orphan genes or genes associated with mobile and extrachromosomal elements, suggestive of probable acquisition by horizontal gene transfer.
Regions of Genomic Plasticity
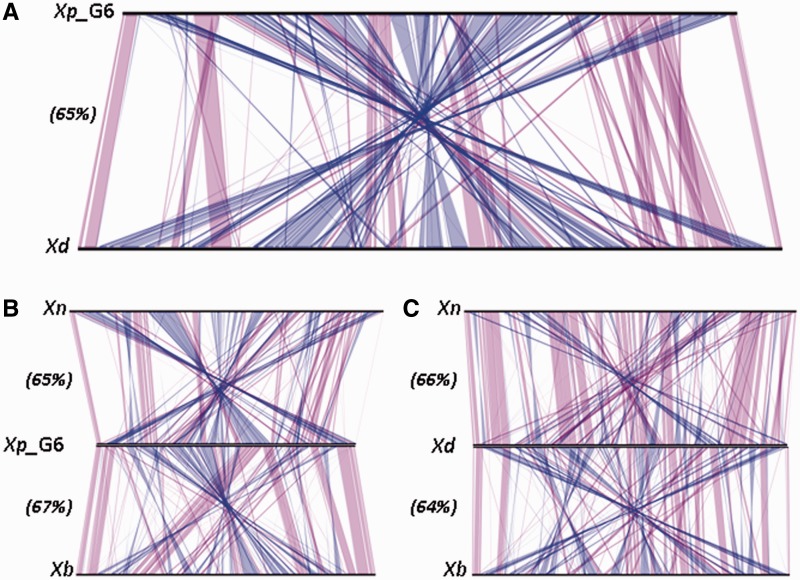
For the vizualization of strain-specific regions, we generated a whole-genome alignment of the sequences of Xp_G6 and Xd (fig. 3A). Despite belonging to closely related species, Xp_G6 and Xd displayed numerous shuffled regions, with synteny conservation for only 65% of the CDS. The large-scale genome rearrangements revealed by synteny comparison were not correlated with differences in genome sizes: whole-genome alignments of Xp_G6 and Xd with Xb and Xn (fig. 3B and C) displayed similar rearrangement patterns and similar percentages of CDS in synteny (64–67%). Genome rearrangements are, therefore, widespread within the genus Xenorhabdus, as previously described for Xb and Xn (Ogier et al. 2010).
Fig. 3.—
Whole-genome sequence alignments between Xenorhabdus genomes. The line plots were obtained with the results for synteny between (A) X. poinarii G6 (Xp_G6) and X. doucetiae FRM16 (Xd); (B) Xp_G6, Xn, and Xb; (C) Xd, Xn, and Xb. Matches between synteny groups occurring on the same strand are shown in purple; matches between synteny groups occuring on the opposite strand are shown in blue. Numbers in brackets indicate the percent of CDS in synteny for each whole-genome alignment.
The large-scale genome rearrangements revealed by genome synteny comparison may result from recombination events (horizontal gene transfer, duplications, inversions, deletions) in the flexible genome. The flexible genome is often structured into RGP, which contain mobile genetic elements, such as genomic islands (GI) and prophage loci, and hypervariable segments, hereafter referred as RGPsensu stricto (Ogier et al. 2010). We identified the RGP of the Xb and Xn genomes by comparing these genomes with a set of Enterobacteriaceae genomes, using the RGPFinder tool (Ogier et al. 2010). In this study, we carried out a new analysis with the four Xenorhabdus genomes as the set of genomes for comparison, leading to the identification of 57, 67, 73 and 79 RGP in the Xp_G6, Xd, Xb and Xn genomes, respectively. Xp_G6 had the smallest number of RGP, accounting for 34% of the entire genome, versus 40–43% for the other three Xenorhabdus genomes considered (table 4 and supplementary table S5, Supplementary Material online). No integral RGP was conserved in all four genomes and, as previously described, only subregions of RGP, named modules, were conserved. This suggests that modules are the true units of plasticity in Xenorhabdus genomes (Ogier et al. 2010).
Table 4.
Number and Classification of RGP in the Xenorhabdus nematophila ATCC19061, X. bovienii SS-2004, X. doucetiae FRM16, and X. poinarii G6 Genomes as a Function of Their Genetic Composition (numbers in brackets indicate the percentage of total genome size)
| Number of Prophages (%) | GI (%) | RGPsensu strictoa (%) | |
|---|---|---|---|
| X. poinarii G6 | 7 (7.3) | 25 (16.4) | 25 (8.8) |
| X. doucetiae FRM16 | 7 (3.7) | 27 (19) | 33 (19.3) |
| X. bovienii SS-2004 | 8 (7.1) | 24 (16) | 41 (14.3) |
| X. nematophila ATCC19061 | 6 (6.2) | 30 (21.5) | 43 (15.6) |
aRGP that are not prophages or GI.
We searched for ICEs among the RGP. By contrast to what has been shown for the closely related strain Photorhabdus luminescens TT01, but similarly with the ICEHIn1056 of Haemophilus influenza strain 1056, the potential Xenorhabdus ICEs consisted of only a remnant of the pilus synthesis locus, an entire or partial pilL gene (supplementary table S6, Supplementary Material online). This part is not essential for ICE self-mobilization (Seth-Smith et al. 2012). Thus, each of the four strains has one entire chromosomal ICE without a pilus synthesis locus and one partial chromosomal ICE, lacking the other features of canonical ICEs. Moreover, Xn harbors an entire ICE with no pilus synthesis locus on its megaplasmid and a partial copy of it on the chromosome. This last feature probably results from integration of the Xn plasmid into the chromosome, followed by gene loss and plasmid immobilization. In addition to ICEs, we also classified RGP into GI, prophages, or RGPsensu stricto. We found similar numbers of GI in Xp_G6, Xd, Xb, and Xn (25, 27, 24, and 30, respectively; table 4). GI were generally located within conserved integration hot spots throughout the genome (supplementary table S5, Supplementary Material online), but gene content was rarely conserved within the GI. The ProPhinder tool allowed us to classify some RGP as prophages (Lima-Mendez et al. 2008; Ogier et al. 2010) (table 4). The four genomes were found to harbor similar numbers of prophage regions. Two P2-related phage clusters have already been described in Xb (xbp1 and xbp2) and Xn (xnp1 and xnp2), and xbp1 and xnp1 have been shown to encode the components of a phage tail-like bacteriocin (Morales-Soto et al. 2012). Likewise, we described xdp1 and xdp2 in Xd and xpp1 in Xp_G6 (supplementary table S5, Supplementary Material online). Finally, RGPsensu stricto, which did not display features of foreign DNA acquired by HGT, accounted for only 9% of the entire Xp_G6 genome, whereas they accounted for 14–19% of the other three genomes (table 4).
Genomic Content of Xp_G6 and Xd and Comparative Analysis with Xb and Xn Genomes
We screened the content of the Xp_G6 and Xd genomes and compared it with that of the Xb and Xn genomes by two approaches. We first searched for genes or loci potentially involved interactions with the host and/or environment on the basis of their annotation. We then systematically searched for genes specifically absent from Xp_G6 and present in the other genomes (supplementary table S7, Supplementary Material online). The genomic regions or genes with a remarkable distribution in the four genomes are listed in table 5.
Table 5.
Selection of Genomic Regions or Genes of Interest Regarding the Bacterial Life-Cycle in the Xenorhabdus poinarii G6 (Xp_G6), X. doucetiae FRM16 (Xd), X. nematophila ATCC19061 (Xn), and X. bovienii SS2004 (Xb) Genomes
| Function | Product or Function | Gene or Locus |
|||
|---|---|---|---|---|---|
| Xn | Xb | Xd | Xp_G6 | ||
| T5SS (Tps) | XhlA/B | XNC1_4556-4555 | XBJ1_0258-0258 | XDD1_3768-3767 | XPG1_0219-0220 |
| Putative CDI | Absent | XBJ1_1975-79 | XDD1_1117-1119 | Absent | |
| Others | XNC1_3685-3689 | Absent | Absent | Absent | |
| XNC1_3564-3565 | |||||
| Hemolysin | XaxAB | XNC1_3766-3767 | XBJ1_1710-1711 | XDD1_0809-0810 | Absent |
| Insecticidal toxin | PirAB | XNC1_1143-1142 | Absent | XDD1_2939-2940 | XPG1_1629-1630 |
| Txp40 | XNC1_1129 | Absent | Absent | Absent | |
| Mcf2 | XNC1_2028 | Absent | XDD1_1049 (truncated) | Absent | |
| Mcf-like | XNC1_2265 | XBJ1_2410 | Absent | Absent | |
| Tc | 3 complete loci, 2 partial locia | 1 complete locus, 4 partial locia | Absent | Absent | |
| NRPS–PKS | PAX-peptide synthesis | XNC1_2781-2784 | XBJ1_2151-2155 | XDD1_2664-2669 | Absent |
| Unknown metabolite synthesis | XNC1_2037-2040 | XBJ1_1966-1968 | XDD1_2281-2289 | Absent | |
| Amino acid-related compounds catabolism | Choline catabolism and transport | XNC1_1244-1247 | XBJ1_3308-3305 | XDD1_1182-1185 | Absent |
| Arginine and amino-butyrate metabolism | XNC1_2270-2274 | XBJ1_0079-0085 | XDD1_1920-1927 | Absent | |
| Hydroxyphenylacetic acid catabolism | XNC1_0446-0449 | XBJ1_3600 | XDD1_0415 | Absent | |
| XNC1_0810-0823 | XBJ1_0867-0875 | XDD1_1012-1026 | Absent | ||
| XBJ1_3599-3560 (partial) | |||||
| Phenylalanine and phenylacetic acid catabolism | XNC1_3619-3621 | XBJ1_3555-3557 | XDD1_0655-0657 | Absent | |
| XNC1_4614-4627 | XBJ1_0117-0128 | XDD1_3917-3928 | Absent | ||
| Tyrosine catabolism | XNC1_2243-2245 | XBJ1_2348-2352 | XDD1_2132-2135 | Absent | |
| Regulators | Two-component system YehU/YehT | XNC1_0512-0513 | XBJ1_0375-0376 | XDD1_0487-0488 | Absent |
| Quorum sensing regulator LuxS | XNC1_1265 | XBJ1_3281 | XDD1_1203 | Absent | |
| Putative RcsA activator | XNC1_1652-1653 | XBJ1_1898 | XDD1_2386 | Absent | |
| Transcriptional repressor for phenylacetic acid degradation | XNC1_4631 | XBJ1_0116 | XDD1_0107 | Absent | |
| Entire TA systems (type IIb) | CcdAB | XNC1_0081-0082 | Absent | Absent | Absent |
| HipBA | XNC1_4231-4232 | Absent | XDD1_0152-0153 | Absent | |
| MazEF | XNC1_0471-0472 | XBJ1_0440-0442 | XDD1_0556-0557, | Absent | |
| XDD1_3836-3837 | |||||
| Doc–Phd | XNC1_0202-0203 | XBJ1_4379-4380 | XDD1_0111-0112 | Absent | |
| RelBE | XNC1_1940-1941 | XBJ1_3187-3188 | Absent | Absent | |
| VapBC | XNC1_0417-0418, | Absent | XDD1_0524-0525, | Absent | |
| XNC1_4632-4633 | XDD1_0577-0578 | ||||
Note.—Tps, two partner system; CDI, contact-dependent inhibitor.
aFor further description, see Chaston et al. (2011).
bPredicted after a RASTA analysis (Sevin and Barloy-Hubler 2007) and from selection of nine families described in Escherichia coli or Yersinia pestis (CcdA/CcdB, HicA/HicB, HigA/HigB, HipA/HipB, MazE/MazF, ParD/ParE, Phd/Doc, RelE/RelB, VapB/VapC).
Secretion Systems
We explored the secretion potential of the four Xenorhabdus strains. The four genomes have nearly similar numbers of T1SS (21, 23, 20 and 18 for Xn, Xb, Xd and Xp_G6, respectively). As previously reported (Chaston et al. 2011), they possess genes encoding the Sec pathway, but they do not encode a T2SS to mediate the crossing of the outer membrane. Unlike the genomes of genus Photorhabdus, Xenorhabdus genomes have no genes encoding a T3SS, confirming the divergence between Xenorhabdus and Photorhabdus in terms of lifestyle (Chaston et al. 2011). We identified two T4SS loci in the Xb, Xd and Xp_G6 genomes and four T4SS loci in the Xn genome, components of entire or partial ICEs (see above). Two T6SS loci were present in the Xn, Xb and Xp_G6 genomes, and Xd was found to carry one additional copy. Finally, the distribution of loci for the T5SSs was particularly marked. The T5SS consists of a transported protein, TpsA, and a channel-forming protein, TpsB, the sole accessory protein devoted to the secretion of TpsA. All four genomes were found to contain a locus encoding XhlA–XhlB, which has been shown to be involved in the export of the XhlA hemolysin responsible for insect virulence in Xn (Cowles and Goodrich-Blair 2005). Only the Xp_G6 genome lacked all the other T5SS systems (table 5).
Insecticidal Toxins, Cytotoxins, and Hemolysins
Xenorhabdus produces an array of insecticidal toxins (Hinchliffe et al. 2010). PirAB (Duchaud et al. 2003; Waterfield et al. 2005) is encoded by the Xn, Xd and Xp_G6 genomes, whereas the Tpx40 toxin (Brown et al. 2006) is encoded by only the Xn genome. None of the Xenorhabdus strains encoded the Mcf1 toxin potentially responsible for triggering apoptosis in insect cells via a BH3-like N-terminal domain (Daborn et al. 2002; Dowling et al. 2007), but the Xp_G6 was the only strain with no Mcf ortholog, Mcf2 or Mcf-like sequence (Waterfield et al. 2003). Interestingly, neither Xd nor Xp_G6 was found to possess loci encoding proteins of the toxin complex (Tc) family (Waterfield et al. 2001). These high-molecular weight proteins have two effects: 1) an oral effect, due to the targeting and disruption of the intestinal epithelium of the lepidopteran Manduca sexta (Bowen et al. 1998) and 2) a phagocytosis-inhibiting effect on insect cells due to the modification of actin and Rho GTPases through ADP-ribosyltransferase activity (Lang et al. 2010; Lango and Clarke 2010). The tc genes are located in GI that were probably acquired by HGT (Waterfield et al. 2002; Ogier et al. 2010). The absence of tc loci from the genomes of Xd and Xp_G6, both of which belong to phylogenetic clade CI (fig. 1), strongly suggests either gene loss or an absence of HGT for tc loci in the bacterial ancestor of phylogenetic clade CI. Interestingly, each Xenorhabdus strain possesses a different cocktail of insecticidal toxins, correlated with phylogenetic status rather than with virulence status. Moreover, the paucity of insecticidal genes in phylogenetic cluster I, which contains both virulent and avirulent strains, argues against a major role for insecticidal toxins in the virulence process.
Xenorhabdus also counteracts cellular immunity by producing cytotoxins and hemolysins (Nielsen-LeRoux et al. 2012). The xaxAB locus of the X. nematophila F1 strain, which encodes a binary pore-forming cytotoxin with apoptotic and necrotic activities in mammalian and insect cells (Vigneux et al. 2007), was absent only from the Xp_G6 genome (table 5). The XaxA and XaxB proteins are probably required for tissue degradation in the cadaver and for efficient subsequent nematode reproduction (Jubelin et al. 2011).
NRPS and PKS
Xenorhabdus protects the insect cadaver from other organisms that might seek to use it as food, by synthesizing an array of secondary metabolites, including antibacterial molecules synthesized by large, multimodular enzymes: the NRPS and PKS (Bode 2009). By searching for NRPS and PKS domains, we identified 16, 13, 12 and 10 loci potentially encoding NRPS/PKS enzymes in the genomes of Xn, Xb, Xd and Xp_G6, respectively. Nevertheless, these similarities in the number of loci conceal considerable differences between the four genomes. Indeed, the number of NRPS/PKS modules, the functional units of the multimodular NRPS–PKS enzymes, was found to be significantly smaller in the Xp_G6 genome (21 modules, 106 kb) than in the Xb, Xd and Xn genomes (56–79 modules, 253–413 kb). Furthermore, the pax locus, encoding NRPS enzymes involved in the synthesis of PAX peptides, which are lysine-rich antifungal cyclolipopeptides (Gualtieri et al. 2009; Fuchs et al. 2011), and an undescribed NRPS locus, weakly similar to a Pseudomonas syringae locus, were specifically absent from the present Xp_G6 genome, but present in the other three Xenorhabdus genomes (table 5). This genomic pattern highlights the low potential of Xp_G6 for the synthesis of secondary metabolites. These metabolites have a wide range of bioactive properties and have been reported to be involved in antimicrobial activities, cytotoxic activity, and immunomodulation in entomopathogenic bacteria (Gualtieri et al. 2009; Park et al. 2009; Vallet-Gely et al. 2010; Fuchs et al. 2011; Stein et al. 2012; Theodore et al. 2012). Their absence from Xp_G6 may limit the capacity of this bacterium to kill the insect on its own.
Catabolism of Amino Acid-Related Compounds
A striking feature of the Xp_G6 genome is the lack of genes encoding proteins involved in the catabolism of amino acid-related compounds: 1) choline and glycine betaine, 2) arginine and amino-butyrate, and 3) aromatic amino acid-related metabolites (supplementary table S7, Supplementary Material online, and table 5). Both primary and secondary metabolisms are required for optimal colonization of the nematode by Ph. luminescens and Xn, but not for bacterial virulence in insects (Martens et al. 2005; Orchard and Goodrich-Blair 2005; Lango and Clarke 2010; Easom and Clarke 2012). Further studies should provide new insight into the possible involvement of such metabolic clusters in X. poinarii pathogenicity.
TA Systems
TA (toxin-antitoxin) systems consist of two closely linked genes, encoding a stable toxin and a labile antitoxin. TA systems are involved in stabilizing genomic regions: when the TA locus is lost, the unstable antitoxin protein disappears first, causing cell death (Van Melderen and De Bast 2009). Additional roles in stress response and/or cell quality control were also recently described (Schuster and Bertram 2013). We identified 42, 12, 37 and 7 genes encoding products with antitoxin or toxin domains in Xn, Xb, Xd and Xp_G6, respectively, but intact TA loci (pairs of colocalized toxin and antitoxin genes) were totally absent from Xp_G6 genome (table 5). Surprisingly, this feature seems to be a general feature of obligate intracellular organisms, whereas free-living slowly growing prokaryotes have a large number of such loci (Pandey and Gerdes 2005).
Small Genome Size and Genomic Reduction, a General Feature of the Species X. poinarii
Small Genome Size
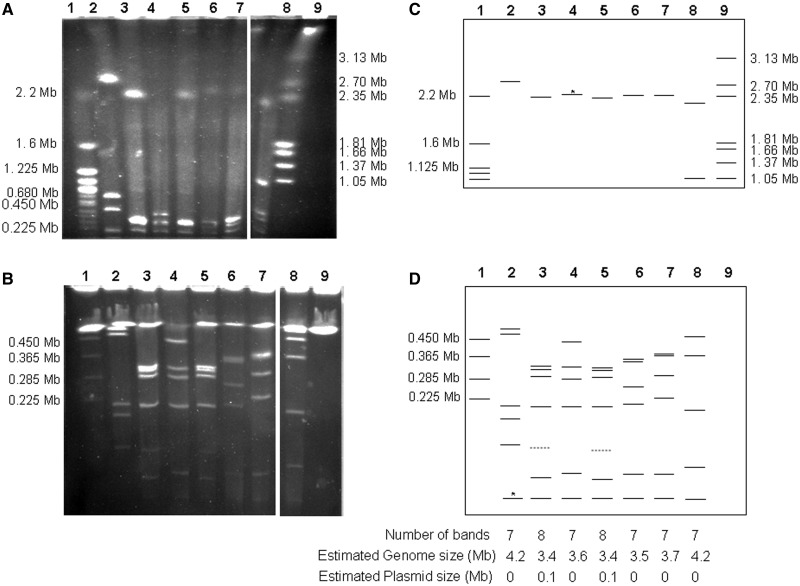
We investigated whether small genome size was a feature particular to the Xp_ G6 strain or a general feature of the species X. poinarii, by examining the whole genome architecture of the other four X. poinarii strains (Xp_SK72, Xp_AZ26, Xp_NC33, and Xp_CU01) by I-CeuI genomic macrorestriction. I-CeuI specifically cleaves the eubacterial 23S rRNA gene of the rrn operon (Liu and Sanderson 1995). Based on the four Xenorhabdus and all the Enterobacteriaceae genome sequences, we expected to obtain seven I-CeuI fragments. The number and sizes of the I-CeuI fragments in the X. poinarii strains were analyzed by PFGE, with migration conditions allowing the separation of fragments from 10 to 4,000 kb (fig. 4). In total, seven DNA bands (ranging from 40 to 2,200 kb in size) were resolved in the gel runs for X. poinarii strains, except for strains Xp_AZ26 and Xp_SK72, for which eight bands were observed. However, the bands of about 120 kb in size obtained for Xp_AZ26 and Xp_SK72 were probably not I-CeuI hydrolysis fragments, corresponding instead to plasmid DNA, given that they were stained less intensely than the other bands (Teyssier et al. 2005). Finally, PFGE analysis of the Xp_SK72, Xp_AZ26, Xp_NC33, and Xp_CU01 strains showed that these strains had genomes ranging in size from 3,400 to 3,700 kb. A small genome is, therefore, a general feature of the species X. poinarii.
Fig. 4.—
Estimation of Xenorhabdus poinarii strains genome size by PFGE of I-CeuI-hydrolyzed genomic DNA. The separation of I-CeuI fragments was optimized by using different electrophoresis conditions for fragments of different sizes: (A) a pulse ramp from 150 to 400 s for 45 h for I-CeuI fragments between 500 and 4,000 kb in size; (B) a pulse ramp from 5 to 35 s for 24 h for fragments of less than 500 kb in size. Schematic representations of the I-CeuI PFGE patterns under two sets of migration conditions, making it possible to separate fragments from 500 to 4,000 kb in size (C) and fragments from 10 to 500 kb in size (D), were also shown. Lane 1: Saccharomyces cerevisiae (strain 972h); lane 2: X. bovienii SS-2004; lane 3: X. poinarii AZ26; lane 4: X. poinarii G6; lane 5: X. poinarii SK72; lane 6: X. poinarii CU01; lane 7: X. poinarii NC33; lane 8: X. doucetiae FRM16; lane 9: Hansenula wingei (strain YB-4662-VIA). Dashed bands around 120 kb in strains Xp_AZ26 (lane 3) and Xp_SK72 (lane 4) correspond to fragments with a lower staining intensity, probably plasmids. *Although these bands are difficult to see on the gel photography, there were directly distinguishable on the gel and their sizes were confirmed by the theorical I-CeuI pattern of the genome sequences of X. bovienii SS-2004 and X. poinarii G6. Fragment and genome sizes of the four unsequenced X. poinarii strains were evaluated with the X. poinarii G6, X. bovienii SS-2004, and X. doucetiae FRM16 genomes used as a reference (lanes 2, 4, and 8) and molecular weight ladders (lanes 1 and 9).
Decay of Isolated Genes
We searched for gene remnants in the Xp_G6 genome, by TBLASTN comparisons of the Xd proteins against the Xp_G6 genome. We found only 24 remnants of Xd genes in Xp_G6 (indicated in supplementary table S7, Supplementary Material online). In Xd, these genes are not clustered together in the same area of the genome. They are instead, spread throughout the genome.
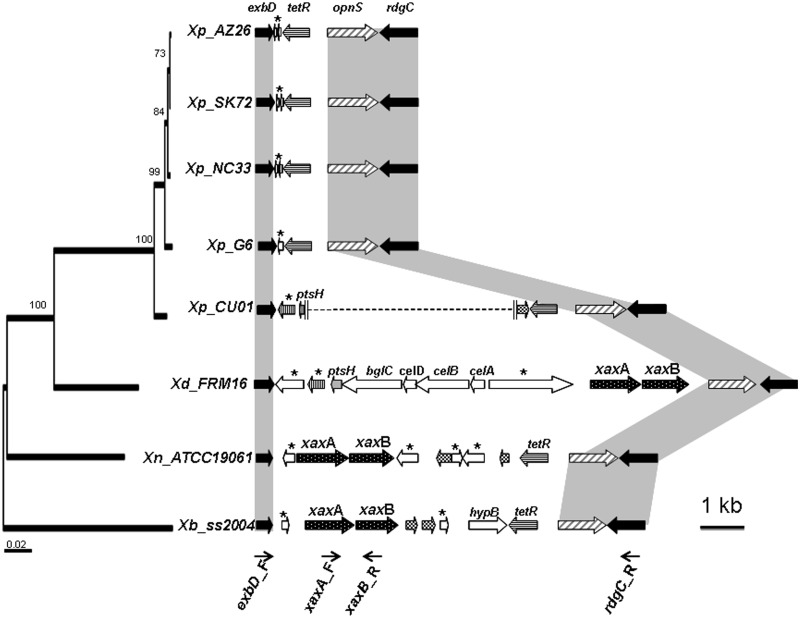
Excisions within RGP: Example of the xaxAB Locus
The xaxAB locus encodes a hemolysin (see above) and is specifically absent from the Xp_G6 strain. In Xd, Xn, and Xb, the xaxAB locus is embedded within RGPsensu stricto (RGP14, RGP64, and RGP28, respectively), a class of RGP specifically underrepresented in the Xp_G6 genome. RGP14, RGP64, and RGP28 are located at the same shuffling point, flanked by the genes of the core genome exbD and rdgC. In the Xp_G6 genome, the genomic content between the exbD and rdgC genes has been significantly reduced, with the presence of only tetR, opnS and one small gene encoding a protein of unknown function (fig. 5). We investigated the presence of xaxAB genes in other X. poinarii strains, by using pairs of primers to amplify the genomic content within the exbD/rdgC shuffling point (exbD_F/rdgC_R). As a control, we first checked that the observed sizes of the amplicons matched the theoretical sizes, for the four sequenced genomes. For Xp_AZ26, Xp_NC33, and Xp_SK72, the size of the amplicon obtained from the sequences between exbD and rdgC was similar to that for Xp_G6 (about 4 kb) and sequencing of the PCR fragments revealed a similar genomic organization in all four strains (fig. 5). Surprisingly, a 10-kb fragment was obtained from the Xp_CU01 genome. Sequencing of the extremities of the regions of the Xp_CU01 amplicon showed the conservation of some Xd genes, with a shuffled genomic organization and the presence of a transposase gene, highlighting progressive genomic erosion in Xp_CU01. The xaxAB locus was not found in the position observed in Xd nor in that observed in Xb/Xn, in Xp_CU01. We checked that the xaxAB locus was not present elsewhere in the X. poinarii genomes, by PCR amplification with the xaxA_F/xaxB_R primer pair on the five X. poinarii strains (data not shown).
Fig. 5.—
The xaxAB locus, its genomic context and its shuffling point exbD/rdgC in the X. doucetiae FRM16 (Xd), X. nematophila ATCC19061 (Xn), X. bovienii SS-2004 (Xb), X. poinarii G6 (Xp_G6), AZ26 (Xp_AZ26), NC33 (Xp_NC33), SK72 (Xp_SK72), and CU01 (Xp_CU01) genomes. The large arrows represent individual ORFs, and the names of the genes are indicated above the arrows. Genes encoding proteins of unknown function are marked with an asterisk. Orthologous genes are indicated by arrows in the same color. Black and chequered arrows represent core-genome genes and transposase genes, respectively. The thin arrows indicate the binding sites of the primers used for PCR amplification. The vertical parallel lines indicate the end of the sequenced area and the dotted lines represent an unsequenced genomic region. The cladogram was obtained by the maximum-likelihood phylogenetic analysis of five concatenated protein-coding sequences (recA, gyrB, dnaN, gltX, and infB), as already described in figure 1. The accession numbers of the sequences of the subsequent amplicons are HG934736 (strain AZ26), HG934737 (strain NC33), HG934738 (strain SK72), HG934739 and HG934740 (strain CU01).
We tested the hypothesis that a deletion event occurred during X. poinarii speciation, by reconstructing the evolutionary history of the xaxAB locus within the Enterobacteriaceae family. We built and compared the topologies of an Enterobacteriaceae phylogenetic tree based on 12 housekeeping genes (fig. 6A) and a xaxA phylogenetic tree (fig. 6B) The Enterobacteriaceae tree grouped the genera into two clades: Providencia–Proteus–Photorhabdus–Xenorhabdus on the one hand and Yersinia–Serratia–Dickeya–Edwarsiella–Erwinia–Klebsiella–Escherichia on the other. We found that xaxA orthologs were present within 1) all the members of the Providencia–Proteus–Photorhabdus–Xenorhabdus clade other than the species Arsenophonus nasoniae and X. poinarii, and 2) only two Yersinia species in the other clade. These results suggest that the xaxA gene was present in the genome of the bacterial ancestor of the Providencia–Proteus–Photorhabdus–Xenorhabdus clade (node A in fig. 6A), from which it was transferred horizontally to the bacterial ancestor of Yersinia kristensenii and Yersinia enterocolitica species (node B in fig. 6A). The most parsimonious hypothesis explaining the absence of xaxA from A. nasoniae and X. poinarii would be the deletion of the locus (crosses in fig. 6A). Arsenophonus nasoniae infects the parasitic wasp Nasonia vitripennis and is responsible for the son-killer trait in wasps (Wilkes et al. 2011). Interestingly, like X. poinarii, A. nasoniae has a significantly smaller (3.6 Mb) genome than its closest relatives, the genera Proteus and Providentia (4–5 Mb), and this genome is not particularly rich in phage genes or transposons (Darby et al. 2010).
Fig. 6.—
Analysis of the evolutionary history of the xaxAB locus by a comparison of topology between an Enterobacteriaceae tree and a xaxA gene tree. (A) Enterobacteriaceae phylogenetic tree based on a maximum-likelihood (ML) analysis of 12 core concatenated protein-coding sequences (infB, nusA, polA, pyrD, rpoB, valS, cysS, metK, purA, tpiA, smpB, secY). Vibrio cholerae sequences were used as the outgroup. Nodes are supported by bootstrap values of more than 93%, unless marked with an asterisk. (B) Phylogenetic tree based on ML analysis of the xaxA gene. Nodes are supported by bootstrap values of more than 86%, unless marked with an asterisk. Node A, the bacterial ancestor of the Providencia–Proteus–Photorhabdus–Xenorhabdus clade, which probably contained the xaxA gene. Node B, bacterial ancestor of the Yersinia kristensenii and Y. enterocolitica species, to which the xaxA gene was probably transferred horizontally. Crosses, probable deletions of the xaxA gene. Vibrio cholerae 16961: NC_002501; Prot. penneri ATCC35198: PRJNA54897; Prot. mirabilis HI4320: NC_010554; Arsenophonus nasoniae DSM15247: PRJNA185551; Prov. stuartii ATCC25827: PRJNA54899; Prov. rettgeri DSM1131: PRJNA55119; Prov. rustigianii DSM 4541: PRJNA55071; Prov. alcalifaciens DSM30120: PRJNA55119; Ph. luminescens TT01: NC_005126.1; Ph. asymbiotica ATCC43949: NC_012962; X. cabanillasii JM26: CBXE010000001-CBXE010000496; X. bovienii SS-2004: NC_013892; X. szentirmaii DSM16638: CBXF010000001-CBXF010000164; X. nematophila ATCC19061: NC_014228.1; X. poinarii G6: FO704551; X. doucetiae FRM16: FO704550; Y. ruckeri ATCC297473: PRJNA55249; Y. pseudotuberculosis IP31758: NC_009708; Y. pestis CO92: NC_003143; Y. intermedia ATCC29909: PRJNA54349; Y. aldovae ATCC35236: PRNJA35243; Y. mollaretii ATCC43969: PRJNA54345; Y. bercovieri ATCC43970: PRJNA54343; Y. rohdei ATCC43380: PRJNA55247; Y. frederiksenii ATCC33641: PRJNA54347; Y. kristensenii ATCC33638: PRJNA55245; Y. enterocolitica 8081: NC_008800; Serratia proteamaculans 568: NC_0098332; Se. odorifera DSM4582: PRJNA40087; Dickeya zeae 1591: NC_012912; Dickeya dadantii 586: NC_013592; Pectobacterium carotovorum PC1: NC_012917; Pe. wasabiae WPP163: NC_013421; Pe. atrosepticum SCRI1043: NC_004547; Edwarsiella tarda EIB202: NC_013508; Edwarsiella ictulari 93-146: NC_012779.2; Pantoea ananatis LMG20103: NC_013956; Erwinia billingiae At-9b: NC_014837; Er. tasmaniensis Eb661: NC_014306; Er. pyrifoliae Ep1/96: NC_02214; Er. amylovora ATCC49946: NC_013971; Klebsiella variicola At-22: NC_013850; K. pneumoniae 342: NC_011283; Salmonella enterica Typhimurium LT2: NC_003197; Sal. enterica Typhi CT18: AL513382; Escherichia albertii TW07627: PRJNA55089; Es. fergusonii ATCC35469T: NC_011740; Es. coli K12: NC_000913).
Discussion
Xenorhabdus bacteria are fascinating models for studies of the mechanisms and evolution of symbioses, because they are both mutualistic symbionts in nematodes and pathogenic symbionts in insects. In recent years, X. nematophila has been widely analyzed, and many molecular and genomic data are now available for this species (Herbert et al. 2007; Nielsen-LeRoux et al. 2012). Several studies have focused on another species, X. bovienii (Chaston et al. 2011, 2013; Kim et al. 2012; Morales-Soto et al. 2012; Sugar et al. 2012). We report here the first analysis of genomic data for the species X. poinarii, which belongs to a phylogenetic group (clade CI), different from that of X. nematophila and X. bovienii. We showed that all the studied strains of X. poinarii had attenuated virulence following their experimental injection into insects. Furthermore, our genomic analysis revealed that a small genome was a general feature of the species X. poinarii. This feature is not typical of the phylogenetic group, because the closely related pathogenic Xenorhabdus strain, Xd, from clade CI (Tailliez et al. 2010), has a genome with a size similar to those of X. nematophila and X. bovienii ones.
The small size of the genomes in the species X. poinarii may reflect either an ancestral state or a recent divergent evolution toward a small genome. In the first hypothesis, all Xenorhabdus strains would have originated from an ancestor with a small genome. In this scenario, X. poinarii would be the only species to have retained a small genome size, with all the others species experiencing genome expansion. However, the evolutionary scenario inferred from the phylogenetic topology based on five genes of the Xcg is not consistent with this hypothesis, because X. poinarii does not occupy a basal position in this phylogeny (fig. 1). According to the second hypothesis, the ancestor of the genus Xenorhabdus had a large genome and deletions have occurred specifically in X. poinarii. We observed both a slight gene decay and a paucity of RGPsensu stricto in X. poinarii (supplementary table S7, Supplementary Material online, and table 4). We thus assume that the RGPsensu stricto (i.e., hypervariable regions of the flexible genome [Ogier et al. 2010]) may have undergone deletion events in the genome of X. poinarii. We therefore propose that the genomes of X. poinarii strains have undergone a reduction with respect to those of other Xenorhabdus genomes, through the excision of genomic blocks from the flexible genome. As an illustration of how such deletion events could occur, we reconstructed the evolutionary history of the xaxAB locus (fig. 6), which is embedded within RGPsensu stricto in the larger genomes of Xn, Xb, and Xd (Ogier et al. 2010). Genomic excision is the most parsimonious hypothesis explaining the absence of the xaxAB locus from the Xp-G6 genome. Several examples of similar deletions have already been reported in bacteria with smaller genomes and weaker virulence than other strains from the same taxon. In Es. coli, the ABU strains have smaller genomes than virulent strains. These strains display frequent point mutations and IS element-mediated deletions in the fim genomic cluster, which is responsible for fimbrial synthesis and the virulence of uropathogenic Es. coli strains (Zdziarski et al. 2008). Likewise, a 77-kb genomic region encoding methionine biosynthesis enzymes, T3SS effectors, and T4SS is deleted in the hypoaggressive Ralstonia solanacearum strain IPO1609. This region contains no features of GI or prophages. Its absence leads to a loss of pathogenicity (Gonzalez et al. 2011).
The compact structure and paucity of nonfunctional sequences in most prokaryotic genomes can generally be accounted for by an inherent deletional bias (Mira et al. 2001; Kuo and Ochman 2009). Host-adapted symbionts (intracellular or niche-restricted) generally have smaller genomes than the free-living bacteria from which they were derived (Moran 2002; Klasson and Andersson 2004). This evolution toward reduced genomes in host-adapted symbionts may be due to genetic drift (Mira et al. 2001; Silva et al. 2001; Nilsson et al. 2005). Indeed, because of sequestration in the host or the occurrence of major lifecycle stages within host, genome of host-adapted bacterial symbionts has reduced opportunity for counterbalancing deletional bias by HGT compared with free-living bacteria. Moreover, the restriction to specific hosts also promotes small bacterial population size, asexuality and population bottleneck for transmission, favoring the persistence of slightly deleterious mutations (Muller’s ratchet). The accumulation of these mutations entails a fitness cost to the bacterium and leads to DNA loss (McCutcheon and Moran 2012). The hallmarks of early stages of genetic drift-driven genomic reduction are an inordinately large number of pseudogenes, a low coding capacity, a high levels of transposable elements, phage-derived sequences, and a massive expansion of IS elements (Toh et al. 2006; Gavotte et al. 2007; Song et al. 2010; Leclercq et al. 2011). Positive selection may also be a significant driver of reductive genome evolution (Koskiniemi et al. 2012). In free-living bacteria growing on a restricted resources in a constant environment, positive selection minimizes the material costs of cellular replication, by reducing genome length (streamlining) (Giovannoni et al. 2005). Large-scale deletions of accessory genes may also be beneficial in a selective environment (Lee and Marx 2012). Comparisons of the large genomes of Xd, Xn, and Xb with the small genome of Xp_G6 highlighted the deletion processes, by revealing large deletions spanning multiple genes (RGPsensu stricto) and small deletions of few nucleotides (gene remnants), together with equivalent coding capacity and the presence of similar numbers of GI, prophages, IS elements, pseudogenes, and phage genes (tables 3 and 4) in the four genomes. The features of the small Xp_G6 genome are therefore rather consistent with a mechanism of selection-driven gene loss in the flexible genome than with a mechanism of genomic reduction dominated by a genetic drift. However, we cannot totally exclude the possibility that genome reduction was also promoted by a population bottleneck. Indeed, as a large proportion of St. glaseri nematodes is naturally aposymbiotic (Akhurst 1986), the transmission of X. poinarii to the next generation of St. glaseri nematodes would involve only a small bacterial population.
The Xenorhabdus lifecycle is characterized by a combination of pathogenic and mutualistic lifestyles and the routine, alternate infection of two kinds of invertebrate hosts. The reduced genome of X. poinarii does not prevent the bacterial/nematode symbiosis from having a lifestyle similar to that of other Xenorhabdus species. What is the evolutionary and ecological significance of genomic reduction in X. poinarii? Selection-driven genome reduction in mutualistic and pathogenic bacteria often results from a greater reliance on the host (Moran et al. 2008). We demonstrated the avirulence of X. poinarii, through direct bacterial injections into two lepidopteran insect species (table 2), as previously reported for several species from the Lepidoptera and Coleoptera (Akhurst 1986; Converse and Grewal 1998; Rosa et al. 2002; Ansari et al. 2003). It is possible that this phylogenetic bacterial group displays greater insect specificity than other Xenorhabdus species. However, no insects susceptible to direct injections of X. poinarii have yet been identified. Alternatively, the bacterial functions necessary for virulence following direct bacterial injection present in other Xenorhabdus species but absent from X. poinarii might be complemented by the nematode partner. Further studies are required to determine the possible role of such complementation in insect virulence.
In conclusion, this first genomic study on the species X. poinarii provides insight into the mechanisms underlying genomic erosion in symbiotic bacteria. In addition, our comparison of the genomes of this avirulent species with those of other Xenorhabdus species paves the way for the identification of new candidate virulence factors in the genus Xenorhabdus.
Supplementary Material
Supplementary tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Nadège Ginibre for assistance with insect pathology assays and Mathieu Sicard for his careful reading of the discussion section. They also thank the INRA MIGALE bioinformatics platform http://migale.jouy.inra.fr for providing computing resources. The paper benefited from the comments of John McCutcheon, and three anonymous reviewers. This study was supported by INRA (grant SPE 2010-1133-01, “Génomique comparative et évolutive de nouveaux facteurs d’adaptation de la bactérie entomopathogène Xenorhadus à ses hôtes insectes”) and by Université Montpellier 2 (grant 2011 “Génomique comparative et fonctionnelle de nouveaux facteurs d’adaptation de la bactérie entomopathogène Xenorhabdus à ses hôtes insectes”).
Literature Cited
- Akhurst RJ. Antibiotic-activity of Xenorhabdus spp, bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ. Taxonomic study of Xenorhabdus, a genus of bacteria symbiotically associated with insect pathogenic nematodes. Int J Syst Bacteriol. 1983;33:38–45. [Google Scholar]
- Akhurst RJ. Xenorhabdus nematophilus subsp poinarii—its interaction with insect pathogenic nematodes. Syst Appl Microbiol. 1986;8:142–147. [Google Scholar]
- Akhurst RJ, Boemare NE. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of Xenorhabdus-Nematophilus to species. J Gen Microbiol. 1988;134:1835–1845. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- Ansari MA, Tirry L, Moens M. Entomopathogenic nematodes and their symbiotic bacteria for the biological control of Hoplia philanthus (Coleoptera : Scarabaeidae) Biol Control. 2003;28:111–117. [Google Scholar]
- Aury JM, et al. High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies. BMC Genomics. 2008;9:603. doi: 10.1186/1471-2164-9-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann BO, Ravel J. Methods for in silico prediction of microbial polyketide and non ribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. Chapter 8. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Bowen D, et al. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- Brown SE, et al. Txp40, a ubiquitous insecticidal toxin protein from Xenorhabdus and Photorhabdus bacteria. Appl Environ Microbiol. 2006;72:1653–1662. doi: 10.1128/AEM.72.2.1653-1662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chaston J, Goodrich-Blair H. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev. 2010;34:41–58. doi: 10.1111/j.1574-6976.2009.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cell Microbiol. 2013;15:1545–1559. doi: 10.1111/cmi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, et al. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One. 2011;6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse V, Grewal PS. Virulence of entomopathogenic nematodes to the western masked chafer Cyclocephala hirta (Coleoptera: Scarabaeidae) J Econ Entomol. 1998;91:428–432. doi: 10.1093/jee/91.2.428. [DOI] [PubMed] [Google Scholar]
- Cowles KN, Goodrich-Blair H. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell Microbiol. 2005;7:899–900. doi: 10.1111/j.1462-5822.2004.00448.x. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Portmann C, Zhang X, Roeffaers MBJ, Clardy J. Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U S A. 2012;109:10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, et al. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc Natl Acad Sci U S A. 2002;99:10742–10747. doi: 10.1073/pnas.102068099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Darby AC, et al. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol Biol. 2010;19:75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling AJ, et al. The Mcf1 toxin induces apoptosis via the mitochondrial pathway and apoptosis is attenuated by mutation of the BH3-like domain. Cell Microbiol. 2007;9:2470–2484. doi: 10.1111/j.1462-5822.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- Duchaud E, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Easom CA, Clarke DJ. HdfR is a regulator in Photorhabdus luminescens that modulates metabolism and symbiosis with the nematode Heterorhabditis. Environ Microbiol. 2012;14:953–966. doi: 10.1111/j.1462-2920.2011.02669.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers RU. Mass production of entomopathogenic nematodes for plant protection. Appl Microbiol Biotechnol. 2001;56:623–633. doi: 10.1007/s002530100711. [DOI] [PubMed] [Google Scholar]
- Ehlers RU, Wulff A, Peters A. Pathogenicity of axenic Steinernema feltiae, Xenorhabdus bovienii, and the bacto-helminthic complex to larvae of Tipula oleracea (Diptera) and Galleria mellonella (Lepidoptera) J Invertebr Pathol. 1997;69:212–217. doi: 10.1006/jipa.1996.4647. [DOI] [PubMed] [Google Scholar]
- Fallon DJ, et al. Effect of entomopathogenic nematodes on Plectrodera scalator (Fabricius) (Coleoptera: Cerambycidae) J Invertebr Pathol. 2006;92:55–57. doi: 10.1016/j.jip.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences—inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fischer-Le Saux M, Arteaga-Hernandez E, Mracek Z, Boemare NE. The bacterial symbiont Xenorhabdus poinarii (Enterobacteriaceae) is harbored by two phylogenetic related host nematodes: the entomopathogenic species Steinernema cubanum and Steinernema glaseri (Nematoda: Steinernematidae) FEMS Microbiol Ecol. 1999;29:149–157. [Google Scholar]
- Fischer-Le Saux M, Viallard V, Brunel B, Normand P, Boemare NE. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp luminescens subsp nov., P. luminescens subsp akhurstii subsp nov., P. luminescens subsp laumondii subsp nov., P. temperata sp nov., P. temperata subsp temperata subsp nov and P. asymbiotica sp nov. Int J Syst Bacteriol. 1999;49:1645–1656. doi: 10.1099/00207713-49-4-1645. [DOI] [PubMed] [Google Scholar]
- Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- Fuchs SW, Proschak A, Jaskolla TW, Karas M, Bode HB. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org Biomol Chem. 2011;9:3130–3132. doi: 10.1039/c1ob05097d. [DOI] [PubMed] [Google Scholar]
- Gaudriault S, et al. Whole-genome comparison between Photorhabdus strains to identify genomic regions involved in the specificity of nematode interaction. J Bacteriol. 2006;188:809–814. doi: 10.1128/JB.188.2.809-814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L, et al. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Rocha EPC, Latorre A, Silva FJ. Reconstructing the ancestor of Mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. 2007;17:1178–1185. doi: 10.1101/gr.6360207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Plener L, Restrepo S, Boucher C, Genin S. Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ Microbiol. 2011;13:3172–3185. doi: 10.1111/j.1462-2920.2011.02636.x. [DOI] [PubMed] [Google Scholar]
- Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- Gualtieri M, Aumelas A, Thaler JO. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J Antibiot. 2009;62:295–302. doi: 10.1038/ja.2009.31. [DOI] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EPC. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han RC, Ehlers RU. Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J Invertebr Pathol. 2000;75:55–58. doi: 10.1006/jipa.1999.4900. [DOI] [PubMed] [Google Scholar]
- Herbert EE, Cowles KN, Goodrich-Blair H. CpxRA regulates mutualism and pathogenesis in Xenorhabdus nematophila. Appl Environ Microbiol. 2007;73:7826–7836. doi: 10.1128/AEM.01586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe SJ, Hares MC, Dowling AJ, Ffrench-Constant RH. Insecticidal toxins from the Photorhabdus and Xenorhabdus bacteria. Open Toxinology J. 2010;3:83–100. [Google Scholar]
- Hussa EA, Goodrich-Blair H. It takes a village: ecological and fitness impacts of multipartite mutualism. Annu Rev Microbiol. 2013;67:161–178. doi: 10.1146/annurev-micro-092412-155723. [DOI] [PubMed] [Google Scholar]
- Jubelin G, et al. Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during insect infection. Environ Microbiol. 2011;13:1271–1284. doi: 10.1111/j.1462-2920.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- Jumas-Bilak E, Michaux-Charachon S, Bourg G, O’Callaghan D, Ramuz M. Differences in chromosome number and genome rearrangements in the genus Brucella. Mol Microbiol. 1998;27:99–106. doi: 10.1046/j.1365-2958.1998.00661.x. [DOI] [PubMed] [Google Scholar]
- Kim SK, Flores-Lara Y, Stock SP. Morphology and ultrastructure of the bacterial receptacle in Steinernema nematodes (Nematoda: Steinernematidae) J Invertebr Pathol. 2012;110:366–374. doi: 10.1016/j.jip.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Klasson L, Andersson SGE. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 2004;12:37–43. doi: 10.1016/j.tim.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Sun S, Berg OG, Andersson DI. Selection-driven gene loss in bacteria. PLoS Genet. 2012;8:e1002787. doi: 10.1371/journal.pgen.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. Deletional bias across the three domains of life. Genome Biol Evol. 2009;1:145–152. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AE, et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- Lango L, Clarke DJ. A metabolic switch is involved in lifestyle decisions in Photorhabdus luminescens. Mol Microbiol. 2010;77:1394–1405. doi: 10.1111/j.1365-2958.2010.07300.x. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Giraud I, Cordaux R. Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol Biol Evol. 2011;28:685–697. doi: 10.1093/molbev/msq238. [DOI] [PubMed] [Google Scholar]
- Lee MC, Marx CJ. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 2012;8:e1002651. doi: 10.1371/journal.pgen.1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLoS Biol. 2003;1:101–109. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- Liu SL, Sanderson KE. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Russell FM, Goodrich-Blair H. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol Microbiol. 2005;58:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nature Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics. 2011;12:116. doi: 10.1186/1471-2105-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Morales-Soto N, Gaudriault S, Ogier JC, Thappeta KRV, Forst S. Comparative analysis of P2-type remnant prophage loci in Xenorhabdus bovienii and Xenorhabdus nematophila required for xenorhabdicin production. FEMS Microbiol Lett. 2012;333:69–76. doi: 10.1111/j.1574-6968.2012.02600.x. [DOI] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nielsen-LeRoux C, Gaudriault S, Ramarao N, Lereclus D, Givaudan A. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr Opin Microbiol. 2012;15:220–231. doi: 10.1016/j.mib.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Nilsson AI, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci U S A. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier JC, et al. Units of plasticity in bacterial genomes: new insight from the comparative genomics of two bacteria interacting with invertebrates, Photorhabdus and Xenorhabdus. BMC Genomics. 2010;11:568. doi: 10.1186/1471-2164-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard SS, Goodrich-Blair H. An encoded N-terminal extension results in low levels of heterologous protein production in Escherichia coli. Microb Cell Fact. 2005;4:22. doi: 10.1186/1475-2859-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, et al. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol Microbiol. 2009;73:938–949. doi: 10.1111/j.1365-2958.2009.06817.x. [DOI] [PubMed] [Google Scholar]
- Penn O, et al. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38:23–28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar GO, Jr, Thomas GM. Significance of Achromobacter nematophilus Poinar and Thomas (Achromobacteraceae: Eubacteriales) in the development of the nematode, DD-136 (Neoaplectana sp. Steinernematidae) Parasitology. 1966;56:385–390. doi: 10.1017/s0031182000070980. [DOI] [PubMed] [Google Scholar]
- Poitout S. Elevage de plusieurs espèces de lépidoptères Noctuidae sur milieu artificiel riche et sur milieu artificiel simplifié. Ann Zool Ecol Anim. 1970;2:79–91. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa JS, Cabral C, Simoes N. Differences between the pathogenic processes induced by Steinernema and Heterorhabditis (Nemata: Rhabditida) in Pseudaletia unipuncta (Insecta: Lepidoptera) J Invertebr Pathol. 2002;80:46–54. doi: 10.1016/s0022-2011(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- Seth-Smith HMB, et al. Structure, diversity, and mobility of the Salmonella pathogenicity island 7 family of integrative and conjugative elements within Enterobacteriaceae. J Bacteriol. 2012;194:1494–1504. doi: 10.1128/JB.06403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin EW, Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard M, et al. Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. Appl Environ Microbiol. 2004;70:6473–6480. doi: 10.1128/AEM.70.11.6473-6480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FJ, Latorre A, Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- Song H, et al. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 2010;6:e1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein ML, et al. One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor. Proc Natl Acad Sci U S A. 2012;109:18367–18371. doi: 10.1073/pnas.1211423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugar DR, et al. Phenotypic variation and host interactions of Xenorhabdus bovienii SS-2004, the entomopathogenic symbiont of Steinernema jollieti nematodes. Environ Microbiol. 2012;14:924–939. doi: 10.1111/j.1462-2920.2011.02663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailliez P, Pagès S, Edgington S, Tymo LM, Buddie AG. Description of Xenorhabdus magdalenensis sp nov., the symbiotic bacterium associated with Steinernema australe. Int J Syst Evol Microbiol. 2012;62:1761–1765. doi: 10.1099/ijs.0.034322-0. [DOI] [PubMed] [Google Scholar]
- Tailliez P, Pagès S, Ginibre N, Boemare N. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int J Syst Evol Microbiol. 2006;56:2805–2818. doi: 10.1099/ijs.0.64287-0. [DOI] [PubMed] [Google Scholar]
- Tailliez P, et al. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis. Int J Syst Evol Microbiol. 2010;60:1921–1937. doi: 10.1099/ijs.0.014308-0. [DOI] [PubMed] [Google Scholar]
- Teyssier C, et al. Pulsed-field gel electrophoresis to study the diversity of whole-genome organization in the genus Ochrobactrum. Electrophoresis. 2005;26:2898–2907. doi: 10.1002/elps.200410323. [DOI] [PubMed] [Google Scholar]
- Theodore CM, King JB, You JL, Cichewicz RH. Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbiotica in liquid media and live crickets. J Nat Prod. 2012;75:2007–2011. doi: 10.1021/np300623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Abraham AL, Touchon M, Rocha EPC. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol Rev. 2009;33:539–571. doi: 10.1111/j.1574-6976.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- Vallenet D, et al. MicroScope-an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41:E636–E647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Opota O, Boniface A, Novikov A, Lemaitre B. A secondary metabolite acting as a signalling molecule controls Pseudomonas entomophila virulence. Cell Microbiol. 2010;12:1666–1679. doi: 10.1111/j.1462-5822.2010.01501.x. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, De Bast MS. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e100043. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrier FJ, Dufort A, Behr MA. The rise and fall of theMycobacterium tuberculosis genome. Trends Microbiol. 2011;19:156–161. doi: 10.1016/j.tim.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Vigneux F, et al. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J Biol Chem. 2007;282:9571–9580. doi: 10.1074/jbc.M604301200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gaugler R, Cui L. Variations in immune response of Popillia japonica and Acheta domesticus to Heterorhabditis bacteriophora and Steinernema species. J Nematol. 1994;26:11–18. [PMC free article] [PubMed] [Google Scholar]
- Waterfield N, Kamita SG, Hammock BD, Ffrench-Constant R. The Photorhabdus Pir toxins are similar to a developmentally regulated insect protein but show no juvenile hormone esterase activity. FEMS Microbiol Lett. 2005;245:47–52. doi: 10.1016/j.femsle.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, Ffrench-Constant RH. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 2001;9:185–191. doi: 10.1016/s0966-842x(01)01978-3. [DOI] [PubMed] [Google Scholar]
- Waterfield NR, Daborn PJ, Ffrench-Constant RH. Genomic islands in Photorhabdus. Trends Microbiol. 2002;10:541–545. doi: 10.1016/s0966-842x(02)02463-0. [DOI] [PubMed] [Google Scholar]
- Waterfield NR, et al. The insecticidal toxin makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol Lett. 2003;229:265–270. doi: 10.1016/S0378-1097(03)00846-2. [DOI] [PubMed] [Google Scholar]
- Wilkes TE, et al. The genus Arsenophonus. In: Zchori-Fein E, Bourtzis K, editors. Manipulative tenants. Boca Raton (FL): CRC Press; 2011. pp. 225–244. [Google Scholar]
- Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect Immun. 2008;76:695–703. doi: 10.1128/IAI.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.