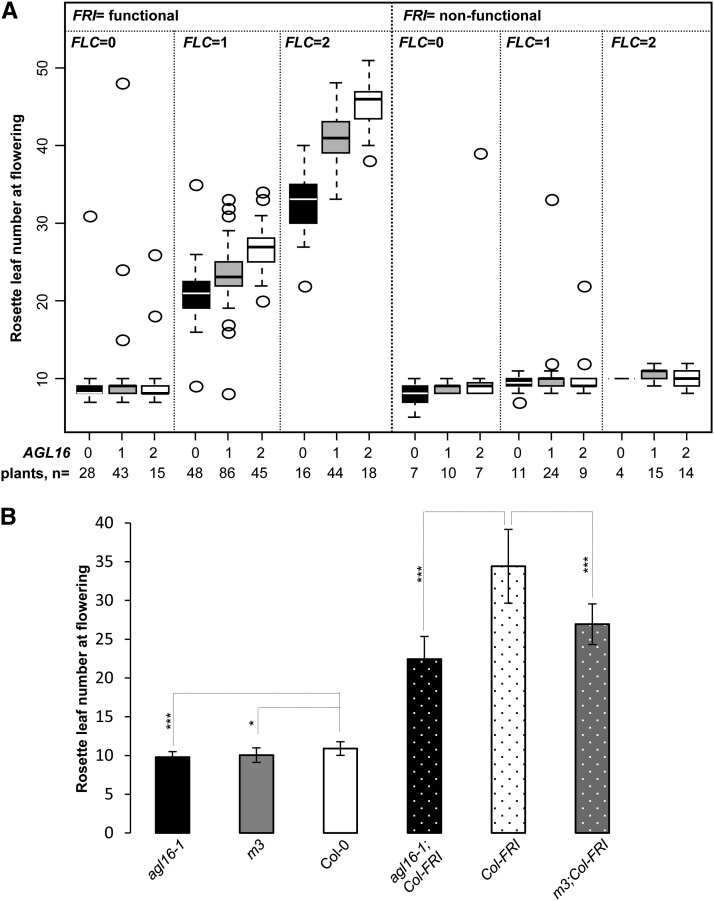
Figure 3.
Flowering Time Is Sensitive to the Quantitative Balance of FLC and AGL16.
(A) RLNs upon flowering are shown for plants of a segregating F2 population of 451 individuals grown under LD conditions. The RLN for each genotype is shown by box plots. Each box encloses the median 50% of the distribution, with the horizontal line marking the median. The lines extending from each box mark the minimum (5%) and maximum (95%) values of the data set. Circles marked the outliers (outside of the 5 to 95% distribution). FRI genotypes are grouped into functional and nonfunctional, as functional FRI is dominant. The allelic numbers of both FLC and AGL16 are coded as 0, 1, and 2, indicating homozygote for knockout allele, heterozygote, and wild-type homozygote, respectively. Numbers at the bottom give the number of plants featuring each multilocus genotype. The AGL16 genotypes alone (P = 0.023) and in interaction with FRI/FLC allelic combinations (P = 0.035) had a significant influence on the flowering time (see main text for details).
(B) Flowering time of plants overexpressing miR824 in both Col-0 (m3) and Col-FRI (m3;Col-FRI) backgrounds. Mean values of rosette leaf number upon flowering for each line together with the sd around the mean were plotted. Differences between lines were evaluated with two-tailed Student’s t test with Bonferroni correction (Supplemental Table 1). ***P < 0.001; *P < 0.05.