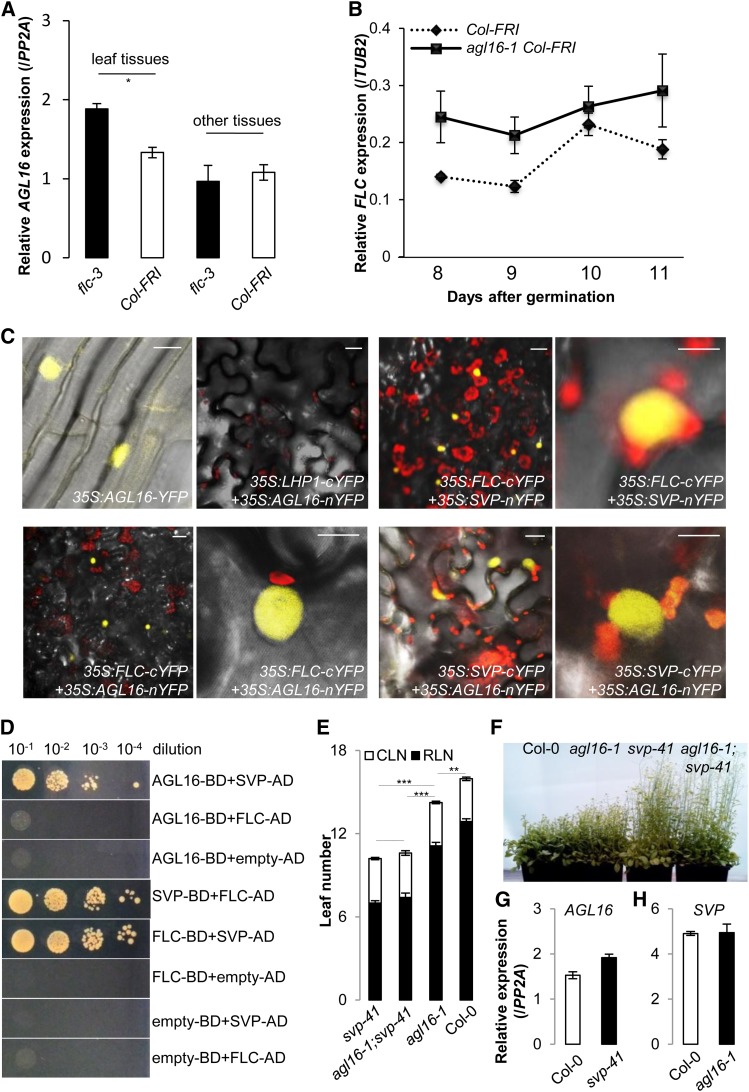
Figure 6.
AGL16 Can Participate in the Repressor Protein Complexes Formed by FLC/SVP.
(A) Loss of function of FLC (flc-3) moderately altered the expression of AGL16 in the Col-FRI background (P < 0.05). Relative accumulation of AGL16 in the flc-3 mutant was monitored by real-time RT-PCR (normalized to PP2A) in different tissues of the aerial parts of 12-d-old seedlings grown under LD conditions. Error bars indicate se of three biological replicates. Note that AGL16 expression in the flc-3 mutant was slightly enhanced compared with Col-FRI (P = 0.02, indicated by one asterisk; two-tailed Student‘s t test). The same pattern was observed in a second independent trial (Supplemental Figure 6A).
(B) Loss of function of AGL16 (agl16-1 Col-FRI) mildly altered the expression of FLC in the Col-FRI background. Relative accumulation of FLC in the agl16-1 Col-FRI mutant was monitored by real-time RT-PCR (normalized to TUB2) along developmental stages from 8 to 11 d after germination on soil under LD conditions. Error bars indicate sd of three technical replicates. The experiment was also performed in the Col-0 background and gave the same pattern (Supplemental Figure 6B).
(C) BiFC assay shows that AGL16 can form a heterodimer with both FLC and SVP in N. benthamiana leaf epidermis. From left to right, top panel, (1) AGL16 is localized in the nucleus, (2) no signal, showing that AGL16 and LHP1 do not interact, (3) positive signal, showing that FLC and SVP interact, and (4) enlarged image of (3). From left to right, bottom panel, (1) positive signal showing that FLC and AGL16 interact, (2) enlargement of (1), (3) positive signal showing that SVP and AGL16 interact, and (4) enlargement of (3). Bars = 10 µm.
(D) Yeast two-hybrid assay confirms the direct interaction between AGL16 and SVP but not between AGL16 and FLC. Each protein was fused to either the activation domain (AD) as prey or DNA binding domain (BD) as bait. Serial dilutions (from 10−1 × to 10−4 ×) of J69-4A cells containing different construct combinations indicated on the right were grown on selective medium. The FLC/SVP combination and the protein/empty vector combinations provide positive and negative controls, respectively.
(E) and (F) Genetic analysis reveals that SVP is epistatic to AGL16. The rosette (filled bars) and cauline (open bars) leaves upon flowering (LD growing) are shown for svp-41, agl16-1 svp-41, agl16-1, and Col-0 wild type. Error bars indicate sd of means. Significance level was tested with the Wilcoxon test (***P < 0.001; **P < 0.01).
(G) and (H) Expression of either AGL16 (G) or SVP (H) is not significantly changed in the loss-of-function mutant svp-41 (G) or agl16-1 (H) compared with Col-0 wild type (normalized to PP2A). Error bars indicate sd of means for three biological replicates.