Abstract
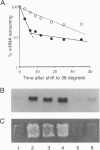
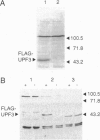
Loss of function of any one of three UPF genes prevents the accelerated decay of nonsense mRNAs in Saccharomyces cerevisiae. We report the identification and DNA sequence of UPF3, which is present in one nonessential copy on chromosome VII. Upf3 contains three putative nuclear localization signal sequences, suggesting that it may be located in a different compartment than the cytoplasmic Upf1 protein. Epitope-tagged Upf3 (FLAG-Upf3) does not cofractionate with polyribosomes or 80S ribosomal particles. Double disruptions of UPF1 and UPF3 affect nonsense mRNA decay in a manner indistinguishable from single disruptions. These results suggest that the Upf proteins perform related functions in a common pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkin A. L., Altamura N., Leeds P., Culbertson M. R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995 May;6(5):611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S. J., Benz E. J., Jr Beta-globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2935–2939. doi: 10.1073/pnas.89.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng J., Maquat L. E. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):482–486. doi: 10.1073/pnas.90.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Cui Y., Hagan K. W., Zhang S., Peltz S. W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995 Feb 15;9(4):423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991 Dec;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the Group II his4-206 and leu 2-3 frameshift mutations. Genetics. 1982 Jul-Aug;101(3-4):345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995 Feb 15;9(4):437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989 Aug 25;17(16):6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989 Oct;123(2):301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hutchison H. T., Hartwell L. H., McLaughlin C. S. Temperature-sensitive yeast mutant defective in ribonucleic acid production. J Bacteriol. 1969 Sep;99(3):807–814. doi: 10.1128/jb.99.3.807-814.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz S. W., Jacobson A., Culbertson M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P., Wood J. M., Lee B. S., Culbertson M. R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992 May;12(5):2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A. J., Olson M. V. Physical map of the Saccharomyces cerevisiae genome at 110-kilobase resolution. Genetics. 1991 Apr;127(4):681–698. doi: 10.1093/genetics/127.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev. 1994 Apr 1;8(7):855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994 Aug 18;370(6490):578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Peltz S. W., Brown A. H., Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993 Sep;7(9):1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Pulak R., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993 Oct;7(10):1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D., Culbertson M. R. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991 May;11(5):2629–2640. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]